Introduction
Human renal cell carcinoma (RCC) is the most common
type of malignant kidney tumor in adults worldwide, and ~85% of
RCCs are clear cell RCC (CCRCC) (1).
RCC is regarded as a localized disease in the early stages,
however, 30% of patients with RCC that present with localized
disease at the time of diagnosis develop metastatic disease within
three years (2). Furthermore, the
prognosis for metastatic RCC is poor (3) as RCC is resistant to traditional
chemotherapy (4,5) and alternative therapeutic strategies for
advanced RCC are limited. At present, novel strategies for the
treatment of advanced RCC include molecular targeted therapy
(6), monoclonal antibodies (7), immunotherapy (8) and the suppression of signaling pathways
(9). Although specific markers
predicting the prognosis of advanced RCC and its potential
therapeutic response to treatment have been investigated, the
molecular mechanisms underlying the progression and development of
RCC have remained elusive.
The forkhead box (FOX) family comprises numerous
proteins with a wide spectrum of biological processes, including
differentiation, metabolism, apoptosis, proliferation, migration
and invasion. FOX proteins contain conserved transcriptional
factors defined by a common DNA-binding domain (10). Furthermore, the FOX family is divided
into 19 subclasses and consists of 50 genes in the human genome
(11). Previous studies have
determined that FOX proteins are associated with carcinogenesis and
the progression of malignancies. For example, the expression of
FOXM1 was increased in a variety of types of tumor, including basal
cell and hepatocellular carcinoma, as well as lung, breast,
prostate and colorectal cancer (12–17). FOXM1
may be associated with carcinogenesis due to its role as a key
regulator in the G1/S and G2/M phases of the cell cycle (18–21). In
addition, FOXO has been reported to be dysregulated in various
types of tumor, including prostate and breast cancer, leukemia,
glioblastoma and endometrial carcinoma (22–28); FOXA1
was overexpressed in thyroid, lung and esophageal cancer (29,30); and
FOXC2 appears to be a key gene involved in tumor progression and
angiogenesis (31).
FOXJ1 is a transcription factor that is significant
in the central nervous and reproductive systems (32–34).
Previous studies have demonstrated that abnormal expression of
FOXJ1 is associated with autoimmune diseases and certain
inflammatory diseases (35,36). This association appears to be due to
the ability of FOXJ1 to suppress T cell activity, resulting in
spontaneous autoimmunity (37). In
addition, FOXJ1 inhibits the humoral immune response in B cells,
with FOXJ1 deficiency in B cells being associated with germinal
center formation and the development of autoantibodies (38). A previous study proposed that FOXJ1
expression was decreased in breast cancer, thus, functioning as a
tumor suppressor gene (39). However,
FOXJ1 expression was increased in hepatocellular carcinoma and was
associated with poor prognosis. Furthermore, overexpression of
FOXJ1 appears to be involved in proliferation and cell-cycle
progression. In brief, little is known regarding the potential
roles of FOXJ1 in carcinogenesis (40).
The expression of FOXJ1 and its function in human
RCC is unclear. Therefore, the current study aimed to determine the
expression of FOXJ1 in human RCC and its effect on the
proliferative ability of human RCC cells.
Materials and methods
Patients and samples
The current study included 286 patients with RCC
that had undergone radical nephrectomy in the Department of Urology
of the Affiliated Hospital of Yanbian University (Yanji, China)
between April 2002 and March 2003. The histological cell type of
all specimen slices was determined by experienced pathologists and
all samples were diagnosed as conventional CCRCC. The clinical
tumor stages and characteristics were classified according to the
tumor node metastasis (TNM) classification system (41), and the nuclear grade was evaluated
according to the Fuhrman grading system of malignant tumors
(42). RCC tissue samples and
corresponding healthy kidney tissues located at a maximal distance
from the tumor were collected immediately following surgical
resection. The samples were formalin-fixed (Sigma-Aldrich, St.
Louis, MO, USA), dehydrated and paraffin-embedded (Sigma-Aldrich).
All tissue samples were maintained in liquid nitrogen
(Sigma-Aldrich) prior to protein and RNA extraction. The patients
were followed up every three months for a period of 120 months. The
present study was approved by the Ethics Committee of the
Affiliated Hospital of Yanbian University and written consent was
obtained from all patients.
Immunohistochemistry
All paraffin-embedded tissue sections (4 µm) were
deparaffinized in xylene (Sigma-Aldrich) and rehydrated.
Subsequently, endogenous peroxidase activity was blocked by
treatment with 0.4% hydrogen peroxide (Sigma-Aldrich) for 20 min
followed by blocking with rabbit serum (Sigma-Aldrich) for 30 min.
The sections were then incubated with primary FOXJ1 monoclonal
mouse anti-rat antibody (cat. no. sc-53139; 1;1,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 37°C for 1 h. The sections
were washed with Tris buffer prior to incubation with biotinylated
polyclonal goat anti-mouse antibody (cat. no. E0433; 1:2,000; Dako,
Glostrup, Denmark) at 37°C for 2 h. Detection of the antibody
reactions was performed using the standard streptavidin-biotin
complex technique (43). The tissue
sections were immunohistochemically examined under a light
microscope (ZX-117M; Shenzhen Zhongxun Optics Instrument Co., Ltd.,
Shenzhen, China), with FOXJ1 expression semi-quantitatively
determined according to staining intensity (−, negative; +, weak;
++, moderate; and +++, strong).
Western blot analysis
Western blot analysis was performed according to the
manufacturer's instructions. Briefly, total protein was isolated
from the CCRCC and healthy tissue samples using lysis buffer
(Sigma-Aldrich) as previously described (44), and the total protein concentration was
determined using a Bradford dye-binding protein assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Subsequently, 10% SDS-PAGE
(Bio-Rad Laboratories, Inc.) was performed. FOXJ1 monoclonal mouse
anti-rat antibody (cat. no. sc-53139; 1;1,000; Santa Cruz
Biotechnology, Inc.) was applied as the experimental antibody and
anti-β-actin monoclonal mouse anti-human antibody (cat. no. ab6276;
1:5,000; Abcam, Cambridge, UK) was applied as a loading control at
37°C for 2 h. The immune complexes were evaluated using an enhanced
chemiluminescence system (GE Healthcare Life Sciences, Chalfont,
UK).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the CCRCC and healthy
kidney tissues using an illustra™ QuickPrep mRNA purification kit
(GE Healthcare, Life Sciences), according to the manufacturer's
instructions, and RT was performed using a First-Strand
complementary (c)DNA synthesis kit (GE Healthcare Life Sciences).
The PCR conditions were determined according to the manufacturer's
instructions as follows: Denaturation at 95°C for 5 min, annealing
for 30 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 1
min and extension at 72°C for 10 min. RT-qPCR was performed using
LightCycler® FastStart DNA Master SYBR Green I (Roche Diagnostics
GmbH, Indianapolis, USA) and the PCR products were detected by
agarose gel electrophoresis, followed by quantification of the
products using LightCycler (Roche Diagnostics GmbH). The primer
sequences were as follows: FOXJ1 forward,
5′-TCGAGATGGCGGAGAGCTGG-3′ and reverse, 5′-GATCCCAAGAAGGCCCCCAC-3′;
GAPDH forward, 5′-ATCAAGAAGGTGGTGAAGCAG-3′ and reverse,
5′-TGGAGGAGTGGGTGTCGC-3′. All RT-qPCR experimental procedures were
conducted in accordance with Minimum Information for Publication of
Quantitative Real-Time PCR Experiments guidelines (45).
Cell culture
Four RCC cell lines (Caki-1, NC65, ACHN, and A498)
were purchased from the American Type Culture Collection (Manassas,
VA, USA). The RCC cell lines were cultured in complete medium
consisting of RPMI-1640 medium (Gibco Bio-Cult Diagnostics Ltd.,
Glasgow, Scotland, UK) supplemented with 25 mM HEPES, 2 mM
glutamine, 10% heat-inactivated fetal bovine serum, 100 µg/ml
streptomycin, 100 U/ml penicillin and 5% non-essential amino acids
(all obtained from Sigma-Aldrich). All RCC cell lines were
maintained as monolayers in 10-cm petri dishes (Corning Inc.,
Corning, NY, USA) and cultured in an incubator with a humidified
atmosphere of 5% CO2 at 37°C.
RNA interference (RNAi) and
transfection
RNAi
RCC cells were incubated in culture dishes with
complete medium at 37°C until cell confluence reached 30–50%.
Subsequently, the RCC cells were transfected with 50 ng/ml small
interfering (si)RNA oligonucleotides against FOXJ1 using
Lipofectamine® 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA). The siRNA oligonucleotide sequences were designed using
siDirect software (http://sidirect2.rnai.jp). Following incubation for 48
h, FOXJ1 expression was evaluated by RT-PCR.
Transfection
The coding sequence of normal human FOXJ1 was
synthesized by RT-PCR using HK-2 (healthy kidney cell line) cDNA
(American Type Culture Collection) as the substrate. The PCR
products of FOXJ1 were then subcloned into the pcDEF3 vector
(Sigma-Aldrich) as described previously (46). The expression vector containing
full-length FOXJ1 cDNA was stably transfected into the four RCC
cell lines using Lipofectamine 2000. G418 (Sigma-Aldrich) was used
to select RCC cells successfully transfected with FOXJ1, and FOXJ1
expression was evaluated by RT-PCR.
Proliferative ability analysis
The effect of FOXJ1 on the proliferative ability of
RCC cells was analyzed using a WST-1 assay. In brief,
exponentially-growing RCC cells were obtained and seeded into a
96-well microtiter plate. Following incubation for 24, 48 and 72 h,
10 µl WST-1 (Roche Diagnostics GmbH, Penzberg, Germany) was added
to each well, and incubated for an additional 2 h. The absorbance,
which represents the cell count in each well, was examined using a
microculture plate reader (Immunoreader NJ-2000; Japan Intermed
Co., Ltd., Tokyo, Japan) at a wavelength of 450 nm.
RCC xenograft mouse models
Thirty BALB/c nude mice (age, 3–4 weeks; Affiliated
Hospital of North Sichuan Medical College, Nanchong, China) were
randomly divided into two groups (control and FOXJ1 vector groups).
The mice were kept in pathogen-free conditions, at temperatures of
26–28°C and 30–40% humidity and were exposed to 12 h light/dark
cycles with free access to food and water. A total of
4×108 RCC cells were administered via subcutaenous
injection into the lumbar region of each mouse. All mice were
observed continuously for five weeks and the volume of each tumor
was measured once a week. Following five weeks, all mice were
sacrificed under deep anesthesia and the final volume of each tumor
was recorded. Tumor volumes (v) were calculated using the following
formula: v = ab2π / 6, where a is the longest diameter
and b is the longest perpendicular diameter.
Statistical analysis
Statistical calculations were performed using SPSS
software (version 19.0; IBM SPSS, Armonk, NY, USA). All experiments
were performed in triplicate and the results are presented as the
mean ± standard deviation. Statistical significance was determined
using a Student's t-test, and the χ2 test was performed
to analyze the association between FOXJ1 expression and
clinicopathological characteristics. In addition, survival curves
were plotted using Kaplan-Meier analysis. P≤0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
The present cohort included 192 male and 94 female
patients (age range, 51–84 years; median age, 67 years), with a
tumor diameter of 1–17 cm (median size, 4.6 cm). The TNM staging
distribution was as follows: Stage I, 147 patients; stage II, 73
patients; stage III, 41 patients; and stage IV, 25 patients. In
addition, the Fuhrman staging distribution was as follows: Grade I,
124 patients; grade II, 97 patients; and grade III, 65 patients
(Table I). The presenting symptoms
included hematuria (28 patients), flank pain (36 patients) and
palpable masses (19 patients). RCC was an incidental finding during
the routine examination of 108 patients. Furthermore, laboratory
analysis indicated an elevated erythrocyte sedimentation rate in 64
patients at the time of diagnosis, while thrombocytopenia,
erythrocytosis and anemia existed in four patients each. Forty-nine
patients exhibited one or more concomitant diseases, including
angina, urolithiasis, diabetes mellitus and valvular heart disease;
12 patients with CCRCC had previously been treated with radical
nephrectomy on the contralateral side; and 29 patients exhibited
metastatic CCRCC at the time of diagnosis.
 | Table I.Association between characteristics
of patients with CCRCC and FOXJ1 expression, detected using
quantitative polymerase chain reaction and
immunohistochemistry. |
Table I.
Association between characteristics
of patients with CCRCC and FOXJ1 expression, detected using
quantitative polymerase chain reaction and
immunohistochemistry.
|
|
|
|
| FOXJ1 protein
expression |
|
|---|
|
|
|
|
|
|
|
|---|
| Characteristic | n | FOXJ1 mRNA
expression, mean ± SD | P-value | – | + | ++ | +++ | P-value |
|---|
| Kidney disease
state |
|
| <0.05 |
|
|
|
| <0.05 |
|
CCRCC | 286 | 2.64±0.35 |
| 24 | 61 | 112 | 89 |
|
|
Healthy | 286 | 0.71±0.18 |
| 156 | 97 | 33 | 0 |
|
| Gender |
|
| >0.05 |
|
|
|
| >0.05 |
|
Male | 192 | 2.67±0.25 |
| 16 | 42 | 74 | 60 |
|
|
Female | 94 | 2.58±0.28 |
|
8 | 19 | 38 | 29 |
|
| Age, years |
|
| >0.05 |
|
|
|
| >0.05 |
|
<60 | 159 | 2.67±0.23 |
| 13 | 34 | 62 | 50 |
|
|
≥60 | 127 | 2.61±0.24 |
| 11 | 27 | 50 | 39 |
|
| Tumor size, cm |
|
| <0.05 |
|
|
|
| <0.05 |
| ≤7 | 147 | 2.07±0.22 |
| 18 | 53 | 50 | 26 |
|
|
>7 | 139 | 3.25±0.33 |
|
6 | 8 | 62 | 63 |
|
| Histological
gradea |
|
| <0.05 |
|
|
|
| <0.05 |
| I | 124 | 1.85±0.18 |
| 21 | 42 | 59 | 2 |
|
| II | 97 | 2.76±0.27 |
|
3 | 17 | 35 | 42 |
|
|
III | 65 | 3.98±0.36 |
|
0 | 2 | 18 | 45 |
|
| Tumor stage |
|
| <0.05 |
|
|
|
| <0.05 |
| I | 147 | 2.07±0.22 |
| 18 | 53 | 50 | 26 |
|
| II | 73 | 2.74±0.26 |
|
6 | 7 | 38 | 22 |
|
| III | 41 | 3.47±0.32 |
|
0 | 1 | 17 | 23 |
|
| IV | 25 | 4.35±0.41 |
|
0 | 0 |
7 | 18 |
|
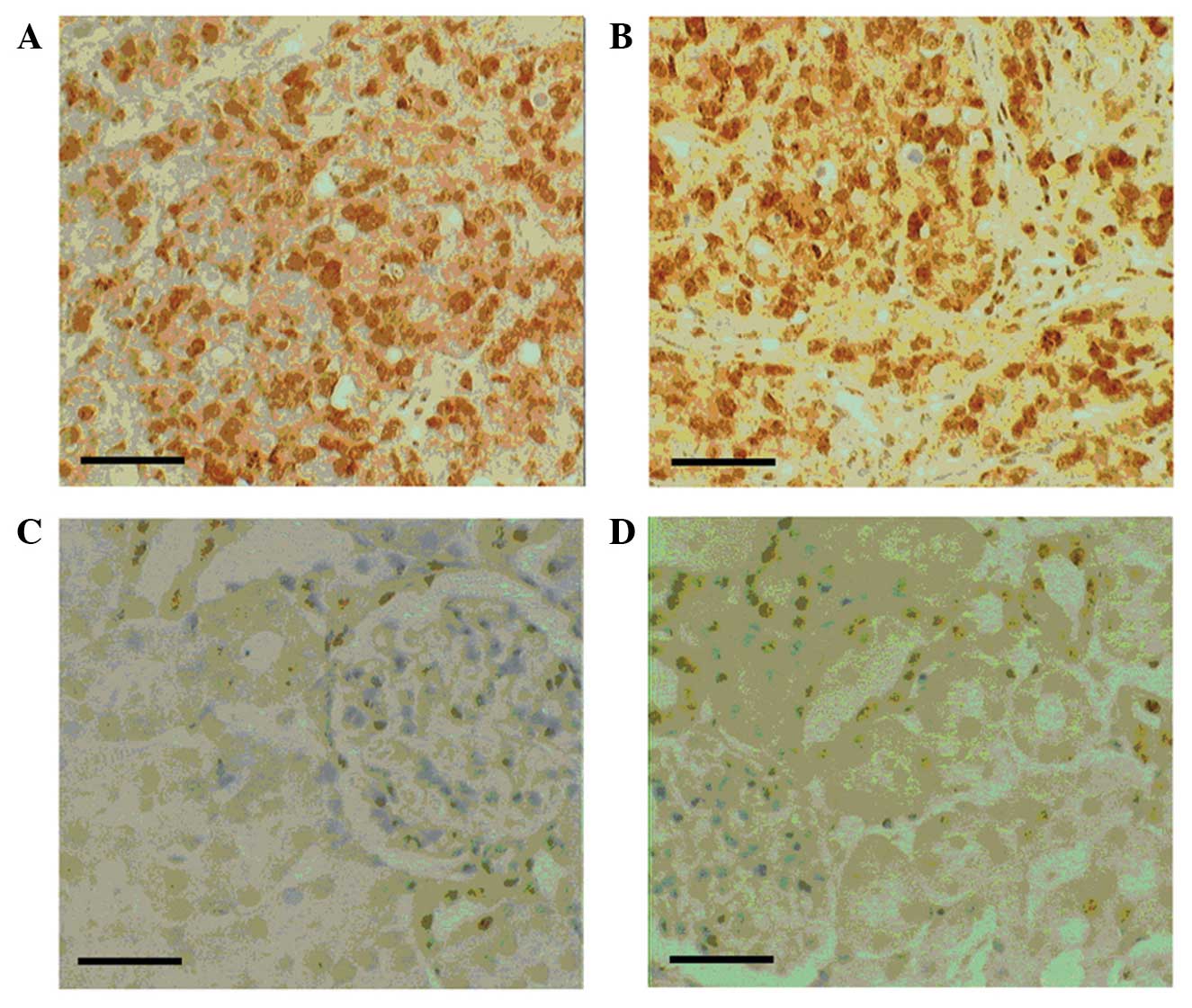
FOXJ1 protein expression in RCC
FOXJ1 protein expression in human CCRCC and healthy
kidney tissues was investigated by immunohistochemical analysis.
FOXJ1 expression appeared to be increased in CCRCC tissues
(Fig. 1A and B) compared with that of
corresponding healthy kidney tissues (Fig. 1C and D). FOXJ1 staining was detected
in the cytoplasm and nuclei of 262/286 CCRCC samples (91.6%), but
in only 130/286 (45.4%) healthy kidney tissue samples. A
significant association was detected between increased FOXJ1
protein expression levels and various clinicopathological
characteristics using χ2 analysis, including advanced
tumor stage, high histological grade and tumor size (P≤0.05).
However, the other investigated characteristics, including gender
and age, did not exhibit a significant association with FOXJ1
protein expression (P>0.05; Table
I). These results indicate that FOXJ1 may be involved in the
carcinogenesis and progression of human CCRCC.
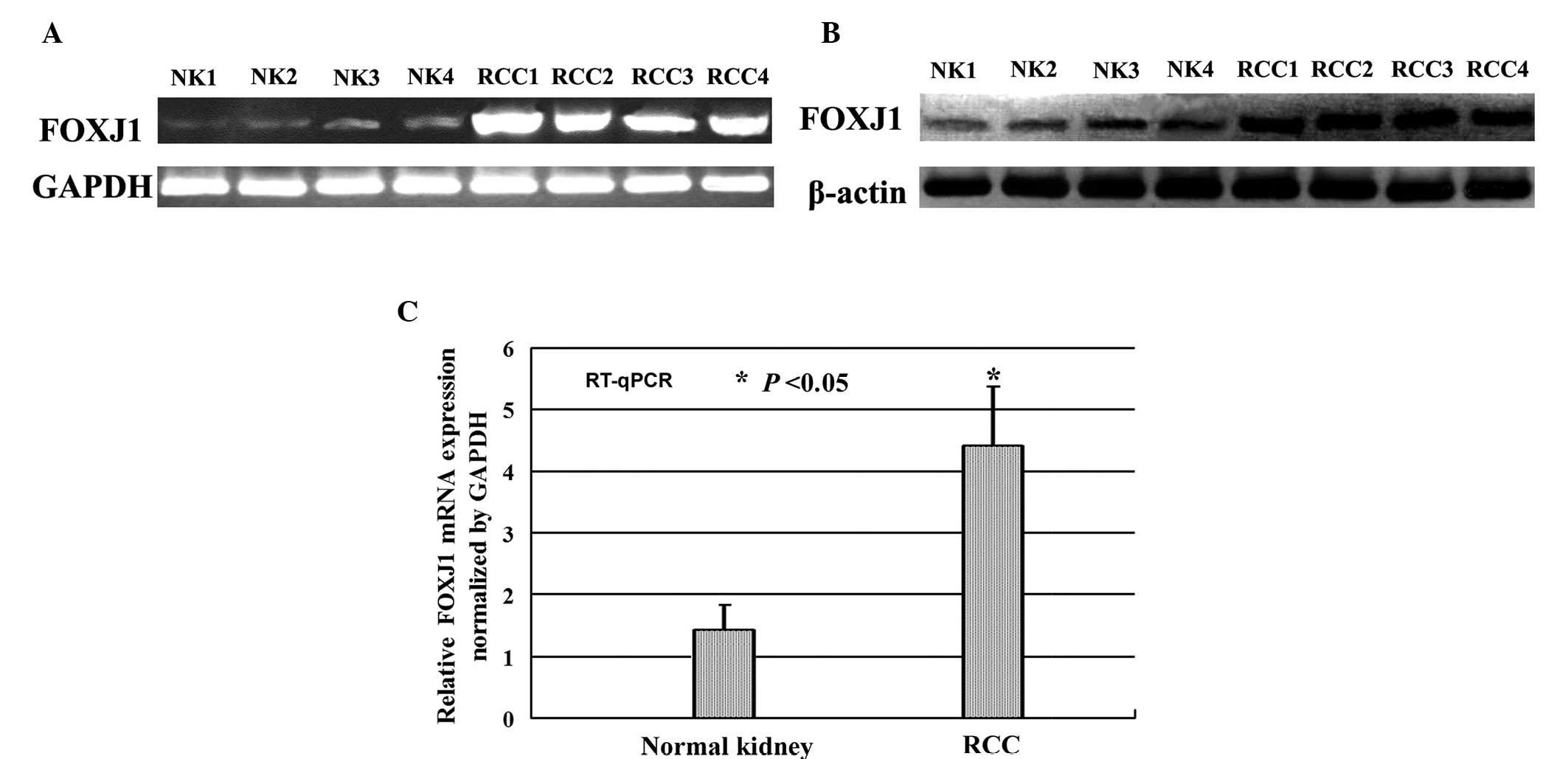
Evaluation of FOXJ1 expression using
RT-PCR, western blotting and RT-qPCR
To clarify the elevated FOXJ1 protein expression
observed in CCRCC by immunohistochemistry, RT-PCR (Fig. 2A) and western blot analysis (Fig. 2B) were performed to determine FOXJ1
expression levels in human CCRCC and healthy kidney tissues. The
relative level of FOXJ1 expression was analyzed by RT-qPCR with
reference to an internal control (Fig.
2C). The results indicated that FOXJ1 expression was
significantly increased in CCRCC tissue compared with that of
corresponding healthy kidney tissues, and FOXJ1 was expressed at
levels similar to those detected by immunohistochemistry. The
results of four pairs of CCRCC and corresponding healthy kidney
tissue samples are indicated in Fig.
2.
Effect of FOXJ1 on the proliferation
of RCC cells
A pcDEF3 vector containing full-length FOXJ1 cDNA
was stably transfected into Caki-1, NC65, ACHN and A498 cell lines.
Additionally, FOXJ1 expression was suppressed using siRNA.
Successful transfections were confirmed using RT-PCR, where FOXJ1
expression was markedly increased by the FOXJ1 vector insert and
markedly decreased by siRNA (Fig.
3A). The effect of FOXJ1 on the proliferation of RCC cells was
determined by performing a WST-1 assay. RCC cells expressing high
levels of FOXJ1 exhibited a significantly increased proliferative
ability compared with that of the control cells. By contrast, RCC
cells expressing low levels of FOXJ1 exhibited lower proliferative
ability compared with that of the control cells (Fig. 3B). The observed increase in
proliferation associated with increased FOXJ1 expression was
supported by identical results obtained from the in vivo
xenograft investigations of BALB/c nude mice (Fig. 3C).
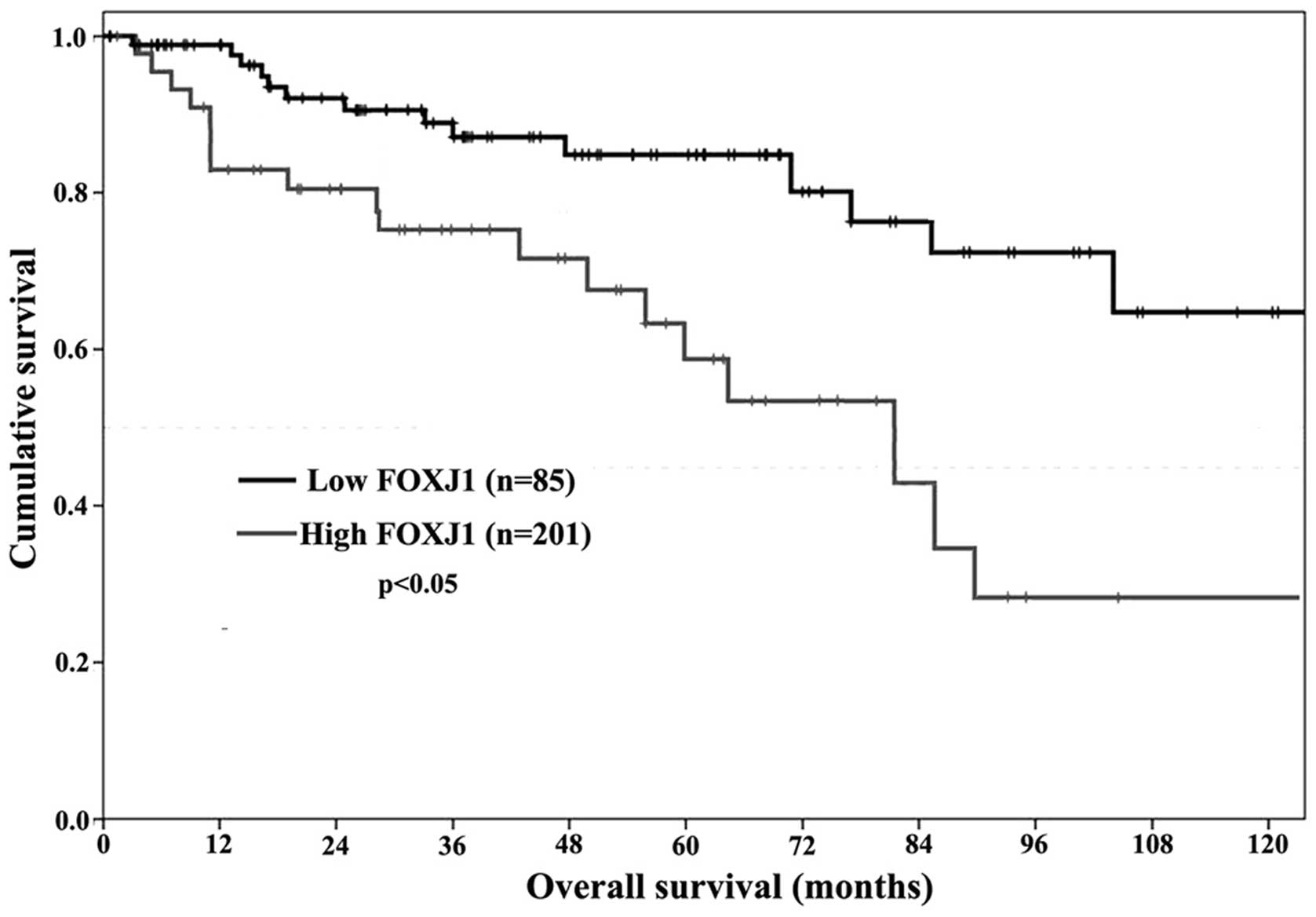
Prognostic significance of FOXJ1
expression
Due to the significant association identified
between FOXJ1 expression, and clinical stage and pathologic grade
in CCRCC, the present study aimed to determine whether FOXJ1 was
able to be regarded as a prognostic marker in human CCRCC. In the
current cohort, 15 patients succumbed to myocardial infarction and
11 patients succumbed to advanced malignant disease. Kaplan-Meier
analysis was performed to calculate the association between FOXJ1
expression and survival in CCRCC. It was demonstrated that the
survival time of patients with CCRCC significantly differed between
the low and high FOXJ1 expression groups (P<0.05; Fig. 4). Furthermore, following 10 years of
follow-up, it was determined that patients expressing
immunohistochemically low levels of FOXJ1 (− and +) lived
significantly longer compared with patients in whom
immunohistochemical staining demonstrated high FOXJ1 expression (++
and +++). These results indicated that FOXJ1 expression may serve
as an independent marker for predicting the prognosis of patients
with CCRCC.
Discussion
Various members of the FOX family, including FOXM1,
FOXO, FOXA1 and FOXC2, have been studied, with the results
indicating that FOX sub-families may be important in the
tumorigenesis and progression of certain carcinomas (10). However, the function of FOXJ1 in
carcinogenesis has remained unclear. To date, the role of FOXJ1 has
generated considerable attention in certain types of tumor, with a
number of studies analyzing its expression in human tumors. For
example, a recent study indicated that FOXJ1 expression was
increased and associated with aggressive characteristics in
hepatocellular carcinoma. Thus, FOXJ1 was proposed as a prognostic
marker in patients with hepatocellular carcinoma (40). By contrast, a previous study proposed
that FOXJ1 was decreased and may function as a tumor suppressor
gene in breast cancer (39). The
expression of FOXJ1 and its role in RCC has remained to be
determined.
To the best of our knowledge, the current study was
the first to investigate FOXJ1 expression in human RCC. FOXJ1
expression levels were determined in human CCRCC samples using
RT-PCR, western blot analysis and RT-qPCR. These methods identified
that FOXJ1 expression levels were similar to those detected by
immunohistochemistry. Additionally, the current study revealed that
FOXJ1 expression was significantly increased in CCRCC compared with
that of healthy kidney tissues. Furthermore, the expression of
FOXJ1 was significantly associated with tumor stage, histological
grade and tumor size. These findings indicated that FOXJ1 may
function as a significant gene that is key in the tumorigenesis and
progression of CCRCC. In vitro and in vivo analysis
of the effect FOXJ1 expression on RCC cell proliferation indicated
that FOXJ1 significantly enhanced the proliferation of RCC cells.
Similar results were detected in xenograft investigations using
BALB/c nude mice. The present study also used Kaplain-Meier
analysis to investigate the association between FOXJ1 expression
and the survival of patients with CCRCC. The results indicated that
high expression of FOXJ1 was associated with poor prognosis in
patients with CCRCC. Thus, it was proposed that FOXJ1 may be
considered as an oncogene and an independent marker for predicting
prognosis in patients with RCC. In addition, the FOXJ1 gene may be
important in the tumorigenesis of renal cancer in adults and high
expression levels of FOXJ1 may accelerate the progression of human
RCC. The effects of FOXJ1 observed in both CCRCC tissues and RCC
cells indicate that the conclusions drawn from these results are
likely to apply to RCC in general. Thus, future studies should
analyze the detailed molecular mechanisms regulated by FOXJ1 in
human RCC.
In conclusion, the current results indicate that
FOXJ1 expression was increased in human RCC and that FOXJ1 enhanced
the proliferation of RCC cells. These findings indicate that FOXJ1
is a significant gene that may be crucial in the tumorigenesis and
progression of human RCC. Thus, silencing of FOXJ1 expression may
present a novel treatment strategy for patients with RCC.
Acknowledgements
The present study was supported by grants from the
China State Scholarship Fund (grant no. 201408220020) and the
Yanbian University Science and Technology Development Item (grant
no. 201259).
References
|
1
|
Deng FM and Melamed J: Histologic variants
of renal cell carcinoma: does tumor type influence outcome? Urol
Clin North Am. 39:119–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Athar U and Gentile TC: Treatment options
for metastatic renal cell carcinoma: a review. Can J Urol.
15:3954–3966. 2008.PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer statistics, 2006. CA Cancer J Clin.
56:106–130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu DS, Chang SY and Ma CP: The expression
of mdr-1-related gp-170 and its correlation with anthracycline
resistance in renal cell carcinoma cell lines and
multidrug-resistant sublines. Br J Urol. 82:544–547. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hartmann JT and Bokemeyer C: Chemotherapy
for renal cell carcinoma. Anticancer Res. 19:1541–1543.
1999.PubMed/NCBI
|
|
6
|
Sciarra A, Gentile V, Salciccia S,
Alfarone A and Di Silverio F: New anti-angiogenic targeted therapy
in advanced renal cell carcinoma (RCC): Current status and future
prospects. Rev Recent Clin Trials. 3:97–103. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dalle S, Thieblemont C, Thomas L and
Dumontet C: Monoclonal antibodies in clinical oncology. Anticancer
Agents Med Chem. 8:523–532. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Coppin C: Immunotherapy for renal cell
cancer in the era of targeted therapy. Expert Rev Anticancer Ther.
8:907–919. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Simpson D and Curran MP: Temsirolimus: In
advanced renal cell carcinoma. Drugs. 68:631–638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jackson BC, Carpenter C, Nebert DW and
Vasiliou V: Update of human and mouse forkhead box (FOX) gene
families. Hum Genomics. 4:345–352. 2010.PubMed/NCBI
|
|
12
|
Wonsey DR and Follettie MT: Loss of the
forkhead transcription factor FoxM1 causes centrosome amplification
and mitotic catastrophe. Cancer Res. 65:5181–5189. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim IM, Ackerson T, Ramakrishna S, et al:
The Forkhead Box m1 transcription factor stimulates the
proliferation of tumor cells during development of lung cancer.
Cancer Res. 66:2153–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee JS, Chu IS, Heo J, et al:
Classification and prediction of survival in hepatocellular
carcinoma by gene expression profiling. Hepatology. 40:667–676.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Teh MT, Wong ST, Neill GW, Ghali LR,
Philpott MP and Quinn AG: FOXM1 is a downstream target of Gli1 in
basal cell carcinomas. Cancer Res. 62:4773–4780. 2002.PubMed/NCBI
|
|
16
|
Kalin TV, Wang IC, Ackerson TJ, et al:
Increased levels of the FoxM1 transcription factor accelerate
development and progression of prostate carcinomas in both TRAMP
and LADY transgenic mice. Cancer Res. 66:1712–1720. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshida Y, Wang IC, Yoder HM, Davidson NO
and Costa RH: The forkhead box M1 transcription factor contributes
to the development and growth of mouse colorectal cancer.
Gastroenterology. 132:1420–1431. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Laoukili J, Kooistra MR, Brás A, et al:
FoxM1 is required for execution of the mitotic programme and
chromosome stability. Nat Cell Biol. 7:126–136. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Costa RH: FoxM1 dances with mitosis. Nat
Cell Biol. 7:108–110. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Kiyokawa H, Dennewitz MB and Costa
RH: The Forkhead Box m1b transcription factor is essential for
hepatocyte DNA replication and mitosis during mouse liver
regeneration. Proc Natl Acad Sci USA. 99:16881–16886. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang IC, Chen YJ, Hughes D, et al:
Forkhead box M1 regulates the transcriptional network of genes
essential for mitotic progression and genes encoding the SCF
(Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 25:10875–10894. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Accili D and Arden KC: FoxOs at the
crossroads of cellular metabolism, differentiation and
transformation. Cell. 117:421–426. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu MC, Lee DF, Xia W, et al: IkappaB
kinase promotes tumorigenesis through inhibition of forkhead
FOXO3a. Cell. 117:225–237. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seoane J, Le HV, Shen L, Anderson SA and
Massagué J: Integration of Smad and forkhead pathways in the
control of neuroepithelial and glioblastoma cell proliferation.
Cell. 117:211–223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Modur V, Nagarajan R, Evers BM and
Milbrandt J: FOXO proteins regulate tumor necrosis factor-related
apoptosis inducing ligand expression. Implications for PTEN
mutation in prostate cancer. J Biol Chem. 277:47928–47937. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parry P, Wei Y and Evans G: Cloning and
characterization of the t(X;11) breakpoint from a leukemic cell
line identify a new member of the forkhead gene family. Genes
Chromosomes Cancer. 11:79–84. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ward EC, Hoekstra AV, Blok LJ, et al: The
regulation and function of the forkhead transcription factor,
Forkhead box O1, is dependent on the progesterone receptor in
endometrial carcinoma. Endocrinology. 149:1942–1950. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Goto T, Takano M, Albergaria A, et al:
Mechanism and functional consequences of loss of FOXO1 expression
in endometrioid endometrial cancer cells. Oncogene. 27:9–19. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nucera C, Eeckhoute J, Finn S, et al:
FOXA1 is a potential oncogene in anaplastic thyroid carcinoma. Clin
Cancer Res. 15:3680–3689. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin L, Miller CT, Contreras JI, et al: The
hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on
chromosome band 14q13 is amplified and overexpressed in esophageal
and lung adenocarcinomas. Cancer Res. 62:5273–5279. 2002.PubMed/NCBI
|
|
31
|
Sano H, Leboeuf JP, Novitskiy SV, et al:
The Foxc2 transcription factor regulates tumor angiogenesis.
Biochem Biophys Res Commun. 392:201–206. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brody SL, Yan XH, Wuerffel MK, Song SK and
Shapiro SD: Ciliogenesis and left-right axis defects in forkhead
factor HFH-4-null mice. Am J Respir Cell Mol Biol. 23:45–51. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hackett BP, Brody SL, Liang M, Zeitz ID,
Bruns LA and Gitlin JD: Primary structure of hepatocyte nuclear
factor/forkhead homologue 4 and characterization of gene expression
in the developing respiratory and reproductive epithelium. Proc
Natl Acad Sci USA. 92:4249–4253. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Clevidence DE, Overdier DG, Tao W, et al:
Identification of nine tissue-specific transcription factors of the
hepatocyte nuclear factor 3/forkhead DNA-binding-domain family.
Proc Natl Acad Sci USA. 90:3948–3952. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li CS, Zhang Q, Lim MK, et al: Association
of FOXJ1 polymorphisms with systemic lupus erythematosus and
rheumatoid arthritis in Korean population. Exp Mol Med. 39:805–811.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li CS, Chae SC, Lee JH, Zhang Q and Chung
HT: Identification of single nucleotide polymorphisms in FOXJ1 and
their association with allergic rhinitis. J Hum Genet. 51:292–297.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Srivatsan S and Peng SL: Cutting edge:
Foxj1 protects against autoimmunity and inhibits thymocyte egress.
J Immunol. 175:7805–7809. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin L, Brody SL and Peng SL: Restraint of
B cell activation by Foxj1-mediated antagonism of NF-kappa B and
IL-6. J Immunol. 175:951–958. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Demircan B, Dyer LM, Gerace M, Lobenhofer
EK, Robertson KD and Brown KD: Comparative epigenomics of human and
mouse mammary tumors. Genes Chromosomes Cancer. 48:83–97. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen HW, Huang XD, Li HC, et al:
Expression of FOXJ1 in hepatocellular carcinoma: correlation with
patients' prognosis and tumor cell proliferation. Mol Carcinog.
52:647–659. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Elmore JM, Kadesky KT, Koeneman KS and
Sagalowsky AI: Reassessment of the 1997 TNM classification system
for renal cell carcinoma. Cancer. 98:2329–2334. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hong SK, Jeong CW, Park JH, Kim HS, Kwak
C, Choe G, Kim HH and Lee SE: Application of simplified Fuhrman
grading system in clear-cell renal cell carcinoma. BJU Int.
107:409–415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abourbih S, Sircar K, Tanguay S, Kassouf
W, Aprikian A, Mansure J and Brimo F: Aldehyde dehydrogenase 1
expression in primary and metastatic renal cell carcinoma: an
immunohistochemistry study. World J Surg Oncol. 11:2982013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dutta KK, Nishinaka Y, Masutani H, et al:
Two distinct mechanisms for loss of thioredoxin-binding protein-2
in oxidative stress-induced renal carcinogenesis. Lab Invest.
85:798–807. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bustin SA, Benes V, Garson JA, et al: The
MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Goldman LA, Cutrone EC, Kotenko SV, Krause
CD and Langer JA: Modifications of vectors pEF-BOS, pcDNA1 and
pcDNA3 result in improved convenience and expression.
Biotechniques. 21:1013–1015. 1996.PubMed/NCBI
|