Introduction
Lung cancer is one of the most common malignancies,
with a 5-year survival rate of patients with non-small-cell lung
cancer (NSCLC) of only 15%. Early diagnosis and timely, appropriate
treatment are crucial for improving the survival rates of patients
with lung cancer (1–3). Therefore, the identification of lung
cancer molecular markers to improve early diagnosis is a research
field that has attracted significant attention over the last few
years. As the exhaled breath condensate (EBC) is directly obtained
from the respiratory tract and lungs, EBC component testing may
reflect the pathological and physiological function of the lungs
and signify airway inflammation, as well as other respiratory
diseases, including lung cancer (4,5). The aim
of the present study was to investigate the suppressor p16 gene
mutation rate in the EBC of patients with NSCLC, determine its
association with the occurrence and development of lung cancer and
assess its clinical value in NSCLC diagnosis.
Patients and methods
Subjects
A total of 58 patients with NSCLC, who underwent
surgery and pathological diagnosis at the Department of Thoracic
Surgery of the Second Affiliated Hospital of Nantong University
(Nantong, China) between March, 2010 and April, 2012, were included
in this study. None of the patients had received radiotherapy and
chemotherapy preoperatively. Of the 58 patients, 36 were male and
22 female, with a mean age of 63.6±7.4 years (range, 48–78 years).
A total of 32 patients were smokers and the remaining 26 were
non-smokers. Of the 58 cases, 32 were squamous cell carcinomas and
the remaining 26 were adenocarcinomas. A total of 20 patients had
poorly differentiated, 26 moderately differentiated and 12
well-differentiated tumors and 20 patients presented with lymph
node metastases. According to the classification standards of the
Union for International Cancer Control in 2009, the cases were
classified as follows: Stage I, 18 patients; stage II, 24 patients;
and stage III/IV, 16 patients. For the control group, 30 healthy
subjects were selected, including 9 men and 21 women, with a mean
age of 59.7±13.1 years (range, 35–79 years).
This study was approved by the Ethics Committee of
the Second Affiliated Hospital of Nantong University and all the
participants signed the informed consent form.
EBC collection
EBC was collected from healthy controls and NSCLC
patients prior to surgery with the EBC collector (HAAK EK20
EcoScreen; Eric Jaeger, Friedberg, Germany). The collector was
pre-cooled for 20 min and the subjects were instructed to clean
their mouths, wear a nose clip and breath quietly while biting the
mouthpart. After 20 min of quiet breathing, the expiratory air was
transformed into a snow-like substance by condensation. The
collection tube was then removed and 1–3 ml of EBC was collected
following melting of the specimen and transferred to a centrifuge
tube to be stored at −80°C.
Genetic testing
Genomic DNA was extracted from EBC using a DNA
extraction kit (QIAamp Circulating Nucleic Acid Kit; Qiagen Co.
Ltd, Hilden, Germany). GAPDH amplification was used as a positive
control during polymerase chain reaction (PCR) amplification of the
p16 gene exons 1 and 2 in EBC samples. The mutations were then
contrasted using DNASTAR software (DNASTAR, Inc., Madison, WI,
USA).
Primer synthesis
The PCR amplification kit was provided by Takara
Biotechnology (Dalian) Co., Ltd. (Dalian, China). The p16 gene
sequence data were retrieved from GenBank (National Center for
Biotechnology Information, Bethesda, MD, USA). With the assistance
of the applied primer design software Premier 5.0 (Premier Biosoft,
Palo Alto, CA, USA), specific primers were designed, which were
synthesized by Shanghai ShengGong Biological Engineering Technology
Service Co., Ltd. (Shanghai, China). The p16 gene exon primer
sequences and parameters are presented in Table I.
 | Table I.p16 gene exon amplification primer
sequences and parameters. |
Table I.
p16 gene exon amplification primer
sequences and parameters.
| Primer name | Primer sequence
(5′-3′) | Fragment length
(bp) | Annealing temperature
(°C) |
|---|
| Exon 1 |
|
|
|
|
Forward |
CAGCATGGAGCCTTCGGCTGA | 216 | 55 |
|
Reverse |
GCGCTACCTGATTCCAATTC |
|
|
| Exon 2 |
|
|
|
|
Forward |
AGCTTCCTTTCCGTCATGC | 264 | 55 |
|
Reverse |
GCCAGGTCCACGGGCAGA |
|
|
| GAPDH |
|
|
|
|
Forward |
GTGAAGGTCGGAGTCAAC | 356 | 56 |
|
Reverse |
GAGATGATGACCCTTTTGGC |
|
|
Statistical analysis
SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA)
was used for statistical analysis. The NSCLC patients were grouped
according to gender, smoking history, pathological type, degree of
differentiation, lymph node metastasis and tumor stage. The
χ2 test or Fisher's exact probability method were used
to compare the differences in mutation rates among the EBC samples.
P<0.05 was considered to indicate statistically significant
differences.
Results
Electrophoresis of PCR amplification
products of EBC DNA
DNA extracted from the EBC of NSCLC patients was
used as the template. The internal gene GAPDH was amplified by PCR
and agarose gel electrophoresis yielded clear GAPDH amplification
product bands. The PCR amplification of the p16 gene exons 1 and 2
and agarose gel electrophoresis also yielded clear bands.
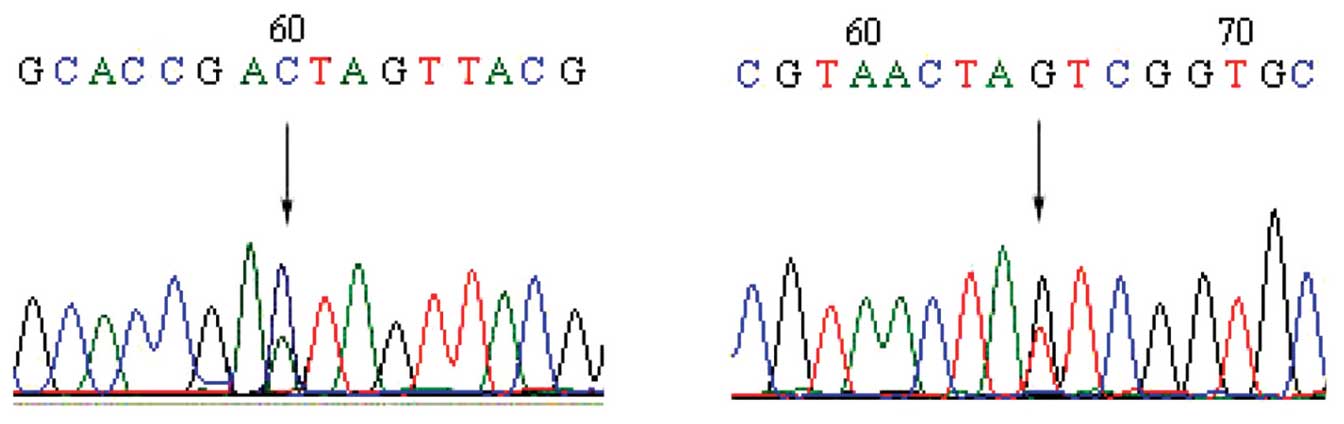
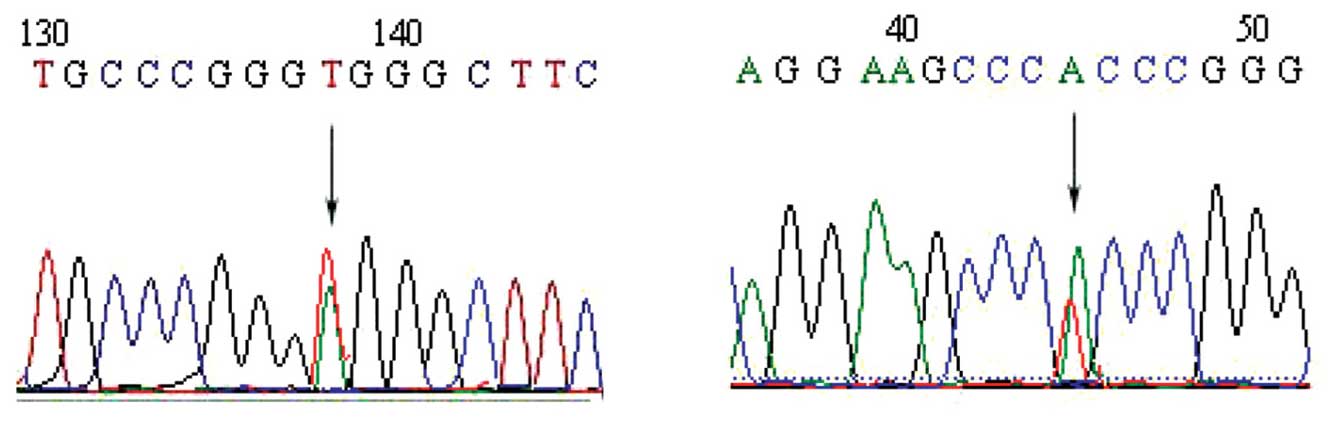
EBC p16 gene sequencing indicates
mutations in NSCLC patients
The direct sequencing method was used to detect p16
gene mutations in the EBC of patients with NSCLC. Of the 58 EBC
specimens from patients with NSCLC, 54 were successfully tested
(the results of 2 cases could not be interpreted and 2 cases did
not conform to the requirements of the test due to a low DNA
concentration). Of the 54 EBC samples, 8 harbored mutations (3 in
exon 1 and 5 in exon 2). Therefore, the mutation rate was 14.81%
(8/54). p16 gene mutations in EBC were not detected in the 30
healthy controls. The p16 gene mutations in the EBC of patients
with NSCLC are presented in Table
II. Sequence diagrams are depicted in Figs. 1 and 2.
 | Table II.p16 gene mutations in the exhaled
breath condensate of patients with non-small-cell lung cancer. |
Table II.
p16 gene mutations in the exhaled
breath condensate of patients with non-small-cell lung cancer.
| Number | Exon | Mutation | Codon | Mutation type |
|---|
| 1 | 1 | GAA→GAC | 94 | Base
substitution |
| 2 | 1 | TGG→AGG | 15 | Base
substitution |
| 3 | 1 | AGC→AGG | 78 | Base
substitution |
| 4 | 2 | GAG→GTG | 88 | Base
substitution |
| 5 | 2 | CCC→CCG | 81 | Base
substitution |
| 6 | 2 | GTG→CTG | 59 | Base
substitution |
| 7 | 2 | GCC→GCG | 86 | Base
substitution |
| 8 | 2 | GAG→GTG | 69 | Base
substitution |
Association between p16 gene mutations
and clinicopathological data of NSCLC patients
EBC p16 gene mutations exhibited no statistically
significant differences according to gender, smoking history,
pathological type, degree of differentiation and presence or
absence of lymph node metastasis. However, EBC p16 gene mutations
exhibited statistically significant differences among different
tumor stages (P<0.05). The p16 gene mutation rate was
proportional to the tumor stage (Table
III). The p16 gene mutation rate in stage I, II and in III/IV
disease was 0.00, 23.53 and 28.57%, respectively.
 | Table III.Association between p16 gene mutations
in the exhaled breath condensate of patients with non-small-cell
lung cancer and clinicopathological characteristics. |
Table III.
Association between p16 gene mutations
in the exhaled breath condensate of patients with non-small-cell
lung cancer and clinicopathological characteristics.
| Clinicopathological
characteristics | p16 gene mutation
cases, n/total (%) | χ2
value | P-value |
|---|
| Gender |
| 1.487 | >0.05 |
| Male | 3/34 (8.82) |
|
|
|
Female | 5/20
(25.00) |
|
|
| Smoking history |
| 0.494 | >0.05 |
| Yes | 6/31
(19.35) |
|
|
| No | 2/23 (8.70) |
|
|
| Pathological
type |
| 0.936 | >0.05 |
| Squamous
cell carcinoma | 3/32 (9.40) |
|
|
|
Adenocarcinoma | 5/22
(22.73) |
|
|
| Degree of
differentiation |
| 3.076 | >0.05 |
|
Well-differentiated | 1/11 (9.09) |
|
|
|
Moderately differentiated | 2/24 (8.33) |
|
|
| Poorly
differentiated | 5/19
(26.32) |
|
|
| Lymph node
metastasis |
| 0.181 | >0.05 |
| Yes | 4/20
(20.00) |
|
|
| No | 4/34
(11.76) |
|
|
| Tumor stage |
| 7.122 | <0.05 |
| I | 0/23 (0.00) |
|
|
| II | 4/17
(23.53) |
|
|
|
III/IV | 4/14
(28.57) |
|
|
Discussion
The prognosis of patients with lung cancer is
closely associated with clinical stage at diagnosis. The 5-year
survival rate postoperatively in patients with stage IA lung cancer
is 60%, while the overall 5-year survival rate of patients with
stage IV cancer is <5%. Early diagnosis and timely, appropriate
treatment are crucial for improving the survival rates of patients
with lung cancer (6). The
identification of alterations in the gene or protein levels
associated with certain types of tumor in the tissues or body
fluids of patients during early tumor formation may significantly
improve early diagnosis of lung cancer.
p16 is a tumor suppressor gene, located in human
chromosome 9, composed of 2 introns and 3 exons. The p16 protein,
encoded by the p16 gene, competes with the cyclin D1 protein for
binding to the cyclin-dependent kinase 4 (CDK4), inhibiting cyclin
D1/CDK4 compound activity and DNA synthesis and ultimately
inhibiting proliferation. Low or no p16 protein expression results
in uncontrollable cell proliferation and tumor growth. Deactivation
of the p16 gene in patients with lung cancer is observed in 25–70%
of the cases (7), suggesting that
alterations of the p16 gene are closely associated with tumor
occurrence.
EBC from the respiratory tract and lung may directly
reflect the pathological and physiological function of the airway.
Data reported by previous studies on lung cancer were mainly
acquired by techniques including detection during surgery,
fiberoptic bronchoscopy biopsy tissue, bronchoalveolar lavage
fluid, bronchoscope brush-off substance, pleural effusion, plasma
and sputum. Compared with the aforementioned methods, EBC detection
technology has the following advantages: i) The EBC collection
process is non-invasive and does not affect airway function or
cause inflammation; ii) it may be repeated within a short period of
time; iii) it may be used as a special detection method for lung
disease specificity; iv) it is a simple, cost-effective and quick
diagnostic method, readily accepted by the patients.
A previous study demonstrated that DNA may be
detected in EBC (8) and Gessner et
al (9) detected four cases of p53
gene mutations among 11 NSCLC patients through EBC collection. Our
previous study also demonstrated that the detection of promoter
methylation of p16 in EBC was feasible (10). These previous studies provide a
theoretical basis for the feasibility of using EBC to detect
genetic changes in patients with lung cancer. The present study
detected p16 gene mutations in the EBC of NSCLC patients and
applied PCR and DNA sequencing to analyze these mutations.
According to the results, the p16 gene mutation rate was 14.81%
(8/54) and p16 gene mutations were not detected in the EBC of
healthy controls, indicating the high specificity of the detection
of gene mutations for the diagnosis of NSCLC. Our results
demonstrated that genetic testing in EBC samples is feasible. The
p16 gene was not detected in certain EBC samples due to the low
concentration of DNA, which did not conform to the concentration
requirements of the sequencing protocol.
When comparing EBC p16 gene mutations according to
gender, smoking history, pathological type, degree of
differentiation and presence or absence of lymph node metastasis,
no statistically significant differences were identified
(P>0.05). However, the p16 gene mutation rate was found to be
proportional to the tumor stage, exhibiting a clear tendency to
increase with advancing stage. This proves that genetic testing is
feasible in EBC samples and that p16 gene mutation detection in EBC
may be used in the evaluation of the condition of the patient.
With the continuous advances in molecular biology,
the application of the knowledge and technology of molecular
biology in the diagnosis of lung cancer and the identification of
novel methods for improving early cancer detection are research
subjects that are attracting increasing attention. EBC appears to
be promising as a novel non-invasive detection method and it may be
used in the diagnosis of lung disease, patient condition
assessment, evaluation of curative effect and prediction of
prognosis. The identification of molecular markers associated with
NSCLC in EBC should be the focus of future studies.
Acknowledgements
This study was supported by a grant from Nantong
Technology Research, Jiangsu Province, China (no. S2006048).
Glossary
Abbreviations
Abbreviations:
|
EBC
|
exhaled breath condensate
|
|
NSCLC
|
non-small-cell lung cancer
|
|
PCR
|
polymerase chain reaction
|
References
|
1
|
Anglim PP, Alonzo TA and Laird-Offringa
IA: DNA methylation-based biomarkers for early detection of
non-small cell lung cancer: an update. Mol Cancer. 7:812008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pillai RN and Ramalingam SS: Advances in
the diagnosis and treatment of non-small cell lung cancer. Mol
Cancer Ther. 13:557–564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koutsokera A, Loukides S, Gourgoulianis KI
and Kostikas K: Biomarkers in the exhaled breath condensate of
healthy adults: mapping the path towards reference values. Curr Med
Chem. 15:620–630. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan HP, Lewis C and Thomas PS: Exhaled
breath analysis: novel approach for early detection of lung cancer.
Lung Cancer. 63:164–168. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mozzoni P, Banda I, Goldoni M, et al:
Plasma and EBC microRNAs as early biomarkers of non-small-cell lung
cancer. Biomarkers. 18:679–686. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Padda SK, Burt BM, Trakul N and Wakelee
HA: Early-stage non-small cell lung cancer: surgery, stereotactic
radiosurgery, and individualized adjuvant therapy. Semin Oncol.
41:40–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Romagosa C, Simonetti S, López-Vicente L,
et al: p16(Ink4a) overexpression in cancer: a tumor suppressor gene
associated with senescence and high-grade tumors. Oncogene.
30:2087–2097. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carpagnano GE, Palladino GP, Gramiccioni
C, et al: New biomolecular methodologies in diagnosis of lung
cancer. Recenti Prog Med. 99:417–421. 2008.(In Italian). PubMed/NCBI
|
|
9
|
Gessner C, Kuhn H, Toepfer K, et al:
Detection of p53 gene mutations in exhaled breath condensate of
non-small cell lung cancer patients. Lung Cancer. 43:215–222. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao P, Chen JR, Zhou F, et al:
Methylation of P16 in exhaled breath condensate for diagnosis of
non-small cell lung cancer. Lung Cancer. 83:56–60. 2014. View Article : Google Scholar : PubMed/NCBI
|