Introduction
HCC is one of the tumors with the highest incidence
worldwide, and its incidence and mortality in China have remained
high. The occurrence of HCC is results from multiple factors,
including the activation of certain oncogenes, inactivation of TSGs
and exogenous stimuli. Previous studies have demonstrated that the
hypermethylation of CpG islands in tumor suppressor gene (TSG)
promoters is closely associated with the formation of HCC, which
may transform the spatial structure of chromatin and hence
block/silence the transcription of TSGs via recruiting proteins of
the methyl CpG binding domain (MBD) family in addition to the
associated protein complexes (1,2).
Epigenetic silencing is a relatively common mechanism of TSG
inactivation (3). The aberrant
methylation of tumor-associated genes, particularly TSGs, requires
further research.
A high frequency of methylation inactivation of the
GSTP1 gene has been observed in human prostate, kidney, and
liver cancers (4–6). Zhang et al (7) and Tchou et al (8) demonstrated that there is a high
frequency of methylation events in the GSTP1 gene in HCC
tumor samples and HCC cell lines, and that the methylation of
GSTP1 in HCC is associated with the action of environmental
carcinogens. As a TSG, P16 gene inactivation may result in
excessive cell proliferation, and the promoter methylation on 9p21
in HCC patients represents the most common mechanism of P16
inactivation (9). Zhong et al
(10) revealed that the abnormal
methylation of the RASSF1A gene promoter is present in 95%
of HCC tissues; the authors hypothesized that the change in
RASSF1A gene expression is an early event during hepatitis B
virus-induced tumorigenesis of HCC.
The present study comparatively analyzed the changes
in methylation level of 4 TSGs in samples from HCC tumors,
tumor-adjacent tissues, and normal liver tissues, and investigated
their correlation with the occurrence and progression of HCC by
consulting the clinicopathological data, in order to provide a
novel way for the early screening and gene diagnosis and therapy of
liver cancer.
Materials and methods
Written informed consent was obtained from the
families of all patients, and the Human Research Ethics Committee
of the Affiliated Nanjing University Drum Tower Hospital (Nanjing,
China) approved the use of all samples under a protocol that
conforms to the provisions of the Declaration of Helsinki (as
revised in Seoul, 2008).
Specimens
The tumor specimens were collected from HCC patients
who had undergone surgical treatment in the Department of
Hepatobiliary Surgery of the Affiliated Drum Tower Hospital, School
of Medicine, Nanjing University, in the Changzhou First People's
Hospital, and in the Third Affiliated Hospital of Soochow
University, during the period between January 2013 and January
2014. The patients did not receive any anticancer treatment and had
no other endocrine, immune, and metabolic diseases prior to the
surgery. Any hemorrhagic and necrotic regions were avoided during
the tumor sample collection. The tumor-adjacent liver tissues were
obtained from the area within 1.5 cm distance from the edge of HCC,
and histologically confirmed for no infiltration of cancer cells.
The normal control group contained 20 cases of pathologically
confirmed normal liver tissue. All the specimens were frozen in
liquid N2 immediately following the resection, then
transported, and stored at −80°C.
Methylation-specific polymerase chain
reaction (MSP) and result interpretation
The DNA samples were extracted from the liver
specimens using a DNA extraction kit (Sangon Biotech Shanghai Co.,
Ltd., Shanghai, China), following the manufacturer's instructions.
The concentration and purity of the extracted DNA were measured on
a UV spectrophotometer (UV-240; Shimadzu Corp., Kyoto, Japan), and
suitable DNA samples were stored at −80°C.
Bisulfite modification of the DNA samples was
conducted using anEZ DNA Methylation-Direct™ Kit (Zymo Research,
Irvine, CA, USA) according to the kit instructions. The DNA samples
were then amplified by MSP and analyzed for methylation status
based on the differential amplification pattern.
The primer sequences, annealing temperature, and
product sizes are presented in Table
I. The PCR system contained PCR Mixture 2X Mix 15 µl, U or
M-Primer F 0.5 µl, U or M-Primer R 0.5 µl, Modified DNA 1–5 µl; the
total volume was adjusted to 30 µl with deionized H2O.
The PCR conditions were as follows: 94°C denaturation, 3 min; 94°C
denaturation 30 sec, annealing 30 sec, and 72°C extension 30 sec,
40 cycles; 72°C extension 7 min. The amplification product was
stored at 4°C. The PCR products (10 µl each) were separated by 2%
agarose gel electrophoresis, and the images were captured on a gel
imaging and analysis system (Yu Long Co., Ltd., Kunming,
China).
 | Table I.Primers for methylation-specific
polymerase chain reaction. |
Table I.
Primers for methylation-specific
polymerase chain reaction.
| Gene | M/U | Primer sequence
(5′-3′) | Annealing
temperature | Product length |
|---|
| GSTP1 | M |
(F)-TTCGGGGTGTAGCGGTCGTC | 61 | 91 |
|
|
|
(R)-GCCCCAATACTAAATCACGACG |
|
|
|
| U |
(F)-GATGTTTGGGGTGTAGTGGTTGTT | 55 | 91 |
|
|
|
(R)-CCACCCCAATACTAAATCACAACA |
|
|
| P16 | M |
(F)-TTATTAGAGGGTGGGGCGGATCGC | 60 | 150 |
|
|
|
(R)-CAACCCCAAACCACAACCATAA |
|
|
|
| U |
(F)-TTATTAGAGGGTGGGGTGGATTGT | 55 | 151 |
|
|
|
(R)-CAACCCCAAACCACAACCATAA |
|
|
| RIZ1 | M |
(F)-GGATTCGCGGTGATTTAC | 69 | 161 |
|
|
|
(R)-AACTCCAATCGAAAAAAACG |
|
|
|
| U |
(F)-ATGGGATTTGTGGTGATTTAT | 58 | 161 |
|
|
|
(R)-CTTAACTCCAATCAAAAAAAACA |
|
|
| RASSF1A | M |
(F)-GGGTTTTGCGAGAGCGC | 52 | 169 |
|
|
|
(R)-CGCTAACAAACGCGAACCG |
|
|
|
| U |
(F)-GGTTTTGTGAGAGTGTGTTTAG | 52 | 169 |
|
|
|
(R)-CACTAACAAACACAAACCAAAC |
|
|
Samples that amplified from the primer pair for
methylated DNA sequence were considered methylation-positive,
whereas samples that amplified from the primer pair for
unmethylated sequence were considered methylation-negative. Samples
with PCR products from both primer pairs were considered partially
methylated, which also represents a methylation-positive
status.
Statistical analysis
The data were statistically analyzed using SPSS
statistical software, version 17.0 (SPSS, Inc., Chicago, IL, USA).
The count and measurement data were compared by χ2 test
and t-test respectively. The association between the gene
methylation in HCC tissues and the clinical data was analyzed using
a Fisher's exact probability test. P<0.05 was used to indicate a
statistically significant difference.
Results
The frequency of methylation of GSTP1 and
RIZ1 genes in HCC tissues were 57.1% (20/35) and 68.6%
(24/35) respectively, both of which were significantly increased
(P<0.01) compared with the levels in adjacent liver tissues
(25.7% [9/35] and 14.3% [5/35] respectively). The frequencies of
methylation of P16 and RASSF1A genes in HCC tissues
were 54.3% (19/35) and 88.6% (31/35), respectively, which did not
significantly differ (P>0.05) from those in adjacent liver
tissues (37.1% [13/35] and 74.3% [26/35]). No methylation of the
GSTP1, P16, and RIZ1 genes was observed in the
20 cases of normal liver tissue, and the methylation of
RASSF1A gene was identified in 10% (2/20) normal liver
tissues (Table II, Figs. 1 and 2).
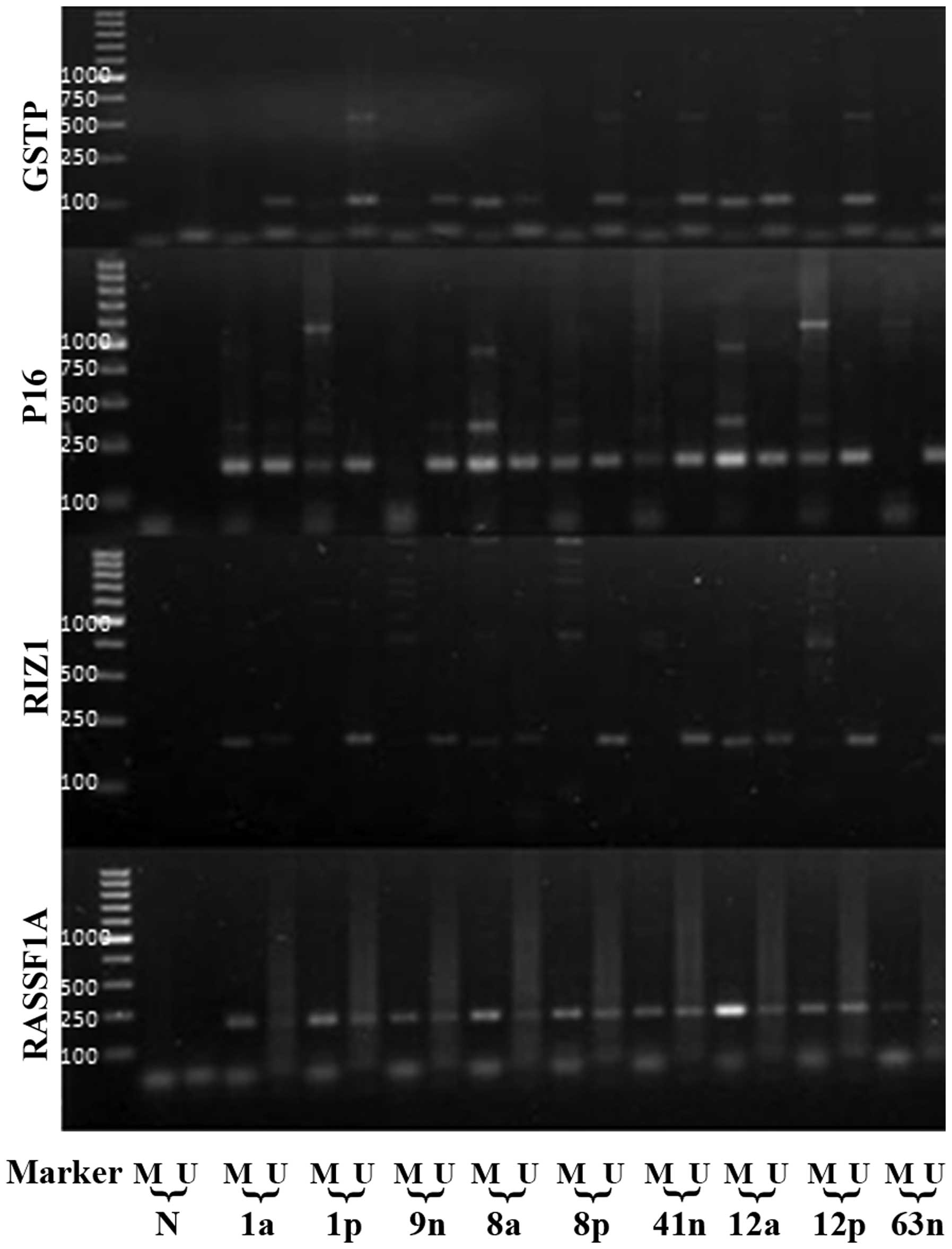 | Figure 2.The MSP products of GSTP1, P16, RIZ1,
and RASSF1A genes in part of the HCC, paired tumor-adjacent, and
normal liver tissues. The lowest marker band represents 100 bp. N,
PCR negative control; M, methylated; and U, unmethylated. a,
cancerous tissue; p, adjacent liver tissue; and n, normal liver
tissue. |
 | Table II.The methylation status of
GSTP1, P16, RIZ1, and RASSF1Agenes. |
Table II.
The methylation status of
GSTP1, P16, RIZ1, and RASSF1Agenes.
|
| Methylation
status |
|---|
|
|
|
|---|
|
| GSTP1 | P16 | RIZ1 | RASSF1A |
|---|
|
|
|
|
|
|
|---|
| Group | + | - | + | - | + | - | + | - |
|---|
| HCC (n=35) | 20 | 15 | 19 | 16 | 24 | 11 | 31 | 4 |
| Adjacent tissue
(n=35) | 9 | 26 | 13 | 22 | 5 | 30 | 26 | 9 |
| Normal liver
(n=20) | 0 | 20 | 0 | 20 | 0 | 20 | 2 | 18 |
| P-value | 0.008a | 0.036b | 0.150a | 0.005b |
<0.01a | 0.199b | 0.124a |
<0.01b |
The frequency of GSTP1 gene methylation in
HCC with capsular invasion was significantly increased compared
with in HCC without capsular invasion (P<0.05). Among the HCC
samples, the frequency of P16 gene methylation in
HbsAg-positive HCC tissues was significantly increased compared
with those in HbsAg-negative HCC tissues (P<0.05). The
methylation status of RIZ1 and RASSF1A genes did not
demonstrate significant correlation with the clinicopathological
data of patients (P>0.05; Tables
III and IV).
 | Table III.The correlation between the
methylation state of GSTP1and P16genes with the
clinicopathological data of HCC patients. |
Table III.
The correlation between the
methylation state of GSTP1and P16genes with the
clinicopathological data of HCC patients.
|
| GSTP1
methylation | P16
methylation |
|---|
|
|
|
|
|---|
|
Clinicopathology | - | + | P-value | - | + | P-value |
|---|
| Gender |
|
| 0.473 |
|
| 0.452 |
|
Male | 11 | 16 |
| 13 | 14 |
|
|
Female | 4 | 4 |
| 3 | 5 |
|
| Age (years) |
|
| 0.281 |
|
| 0.364 |
|
≤50 | 6 | 5 |
| 6 | 5 |
|
|
>50 | 9 | 15 |
| 10 | 14 |
|
| AFP (µg/l) |
|
| 0.518 |
|
| 0.156 |
|
≤20 | 6 | 7 |
| 4 | 9 |
|
|
>20 | 9 | 13 |
| 12 | 10 |
|
| Tumor size
(cm) |
|
| 0.404 |
|
| 0.210 |
| ≤5 | 9 | 10 |
| 7 | 12 |
|
|
>5 | 6 | 10 |
| 9 | 7 |
|
| Cirrhosis |
|
| 0.482 |
|
| 0.347 |
| + | 10 | 12 |
| 9 | 13 |
|
| - | 5 | 8 |
| 7 | 6 |
|
| HbsAg |
|
| 0.668 |
|
| 0.024 |
| + | 12 | 16 |
| 10 | 18 |
|
| - | 3 | 4 |
| 6 | 1 |
|
| Capsular
invasion |
|
| 0.017 |
|
| 0.596 |
| + | 5 | 15 |
| 9 | 11 |
|
| - | 10 | 5 |
| 7 | 8 |
|
| Distal
metastasis |
|
| 0.567 |
|
| 0.481 |
| + | 4 | 6 |
| 4 | 6 |
|
| - | 11 | 14 |
| 12 | 13 |
|
|
Differentiation |
|
| 0.340 |
|
| 0.602 |
| High to
medium | 13 | 15 |
| 13 | 15 |
|
|
Poor | 2 | 5 |
| 3 | 4 |
|
 | Table IV.The correlation of the methylation
state of RIZ1and RASSF1Agenes with the
clinicopathological data of HCC patients. |
Table IV.
The correlation of the methylation
state of RIZ1and RASSF1Agenes with the
clinicopathological data of HCC patients.
|
| RIZ1
methylation | RASSF1A
methylation |
|---|
|
|
|
|
|---|
|
Clinicopathology | - | + | P-value | - | + | P-value |
|---|
| Gender |
|
| 0.492 |
|
| 0.665 |
|
Male | 8 | 19 |
| 3 | 24 |
|
|
Female | 3 | 5 |
| 1 | 7 |
|
| Age (years) |
|
| 0.521 |
|
| 0.372 |
|
≤50 | 3 | 8 |
| 2 | 9 |
|
|
>50 | 8 | 16 |
| 2 | 22 |
|
| AFP (µg/l) |
|
| 0.626 |
|
| 0.522 |
|
≤20 | 4 | 9 |
| 1 | 12 |
|
|
>20 | 7 | 15 |
| 3 | 19 |
|
| Tumor size
(cm) |
|
| 0.352 |
|
| 0.630 |
| ≤5 | 7 | 12 |
| 2 | 17 |
|
|
>5 | 4 | 12 |
| 2 | 14 |
|
| Cirrhosis |
|
| 0.144 |
|
| 0.478 |
| + | 5 | 17 |
| 2 | 20 |
|
| - | 6 | 7 |
| 2 | 11 |
|
| HbsAg |
|
| 0.619 |
|
| 0.609 |
| + | 9 | 19 |
| 3 | 25 |
|
| - | 2 | 5 |
| 1 | 6 |
|
| Capsular
invasion |
|
| 0.281 |
|
| 0.200 |
| + | 5 | 15 |
| 1 | 19 |
|
| - | 6 | 9 |
| 3 | 12 |
|
| Distal
metastasis |
|
| 0.309 |
|
| 0.681 |
| + | 2 | 8 |
| 1 | 9 |
|
| - | 9 | 16 |
| 3 | 22 |
|
|
Differentiation |
|
| 0.619 |
|
| 0.609 |
| High to
medium | 9 | 19 |
| 3 | 25 |
|
|
Poor | 2 | 5 |
| 1 | 6 |
|
Discussion
HCC is one of the tumors with highest incidence
worldwide. Due to an insidious onset, the majority of HCC patients
are diagnosed at an advanced stage, which results in a <20%
clinically resectable rate (11,12). With
the exception of α-fetoprotein (AFP), at present, there remains a
lack of well-recognized effective tumor markers for routine
clinical detection of HCC. Therefore, identifying HCC-associated
tumor markers and studying the underlying molecular mechanisms is
of great importance. Epigenetics refers to a genetic mechanism that
enables the occurrence of inheritable alterations in gene
expression without changing the DNA sequence (3). Epigenetic silencing may result in the
inactivation of TSGs, thereby causing the initiation and
progression of tumorigenesis (3). The
aberrant methylation of tumorigenesis-associated genes,
particularly the TSGs, are receiving more attention.
GSTP1 gene is located on human chromosome
11q13, encoding an enzyme with detoxification and protein-binding
functions (13). Previous studies
have demonstrated that the high-frequency of methylation
inactivation of GSTP1 gene is restricted to certain human
cancers, including prostate, kidney, breast, and liver cancers
(4,5).
Another previous study demonstrated that the frequency of
GSTP1 gene methylation in HCC is 41–85%, and that the
methylation of GSTP1 gene in HCC is associated with the
effect of environmental carcinogens (7). The frequency of GSTP1 gene
methylation in HCC and its cell lines may be as high as ~85%, which
is significantly increased compared with those in abnormal
proliferation-induced liver nodules and cirrhosis tissues (8). The present study demonstrated that the
frequency of GSTP1 methylation in HCC (57.1% [20/35]) was
significantly increased (P<0.01) compared with that in
tumor-adjacent tissues (25.7% [9/35]), while no abnormal
methylation was detected in normal liver tissues. The association
between GSTP1 methylation in HCC and the clinicopathological
data was investigated, and the results demonstrated that the
frequency of GSTP1 methylation in HCC with capsular invasion
was significantly increased compared with that in HCC without
capsular invasion (P<0.05), while no significant correlation
between other clinicopathological features and GSTP1
methylation was detected (P>0.05). Capsular invasion is
associated with the metastasis and invasion of tumor. GSTP1
gene promoter methylation interferes with its normal expression or
function, leading to an accumulation of β-catenin in the cells
(14). Since the latter is important
in mediating the epithelial cell adhesion, such change may
therefore facilitate the intrahepatic metastasis of HCC (6). Therefore, the aberrant methylation of
GSTP1 gene may be associated with the invasiveness of
HCC.
As a TSG, the inactivation of P16 may lead to
excessive cell proliferation, accelerated cell cycle, and hence a
premature entry into the S phase prior to the completion of DNA
repair, resulting in tumorigenesis. Jin et al (9) demonstrated that the promoter methylation
on 9p21 is the most common mechanism for the inactivation of TSG
P16 in HCC. Narimatsu et al (14) studied the methylation status of
P16 in 35 cases of HCC infected by HBV and/or HCV using the
MSP method; their results indicated that P16 methylation may
be induced by hepatitis virus in livers with chronic inflammation
prior to the tumorigenesis of HCC. The present study demonstrated
that the frequency of P16 gene methylation in HCC (54.3%
[19/35]) was increased compared with in the adjacent liver tissues
(37.1% [13/35]), though the difference was not statistically
significant (P>0.05); there was no abnormal methylation of
P16 detected in the normal liver tissues, implying a
potential tumorigenic tendency of the tumor-adjacent tissues and a
possible involvement of P16 methylation in the occurrence of
HCC. Further statistical analysis of the association between the
methylation results and the clinicopathological data of HCC
indicated that the frequency of P16 methylation in
HbsAg-positive patients was significantly increased compared with
in HbsAg-negative ones (P<0.05), indicating that chronic HBV
infection may be the result of methylation inactivation of
P16.
RIZ1 is a TSG that simultaneously regulates
cell proliferation and inhibits oncogenesis (15). The abnormal expression of RIZ1
is associated with the tumorigenesis of a variety of human tumors,
including nasopharyngeal, breast, liver and cervical among other
cancers (16,17). Nomoto et al (18) reported that the frequency of
RIZ1 gene methylation was 45.2% among patients with HCC in
Japan. The results of the present study demonstrated that the
frequency of RIZ1 methylation in HCC (68.6% [24/35]) was
significantly increased (P<0.01) compared with in the paired
adjacent liver tissues (14.3% [5/35]), while no methylation of
RIZ1 was detected in the normal liver tissues. The frequency
of RIZ1 methylation demonstrated a significant gradient
among the 3 groups of samples; the rare presence of RIZ1 in
the adjacent liver tissues indicated highly tumor-specific
epigenetic changes in RIZ1 with the occurrence of HCC,
indicating that the methylation of RIZ1 may be a frequent
event during the tumorigenesis of HCC, and that the appearance of
RIZ1 methylation may indicate an immediate formation of HCC.
Therefore, RIZ1 gene methylation may be of great
significance for the risk assessment and early diagnosis of HCC.
However, further analysis indicated that there was no correlation
between RIZ1 methylation and the clinicopathological data of
HCC patients. It should be noted that since the exact role of
RIZ1 promoter methylation in the process HCC carcinogenesis
remains unclear, further study with an expanded sample size is
required.
RASSF1A is a TSG cloned from lung cancer and
reported by Damman et al in 2000 (19). In certain cases RASSF1A is
inactivated via abnormal promoter methylation in primary HCC. Zhong
et al (10) demonstrated that
the abnormal hypermethylation of the RASSF1A promoter
sequence exists in 95% of HCC tissues, and proposed that
RASSF1A methylation is a relatively early event during the
HBV-induced tumorigenesis of HCC. Previous studies have
demonstrated that the frequency of RASSF1A gene methylation
in HCC is 66.7–100%, and the variation may be associated with
ethnic and geographical variabilities (20,21). In
the present study, 31/35 (88.6%) of HCC specimens presented with
aberrant methylation, as did 26/35 (74.3%)of the corresponding
tumor-adjacent tissues; no statistically significant difference in
aberrant methylation was observed between the above 2 groups
(P>0.05). Taking into account that cancer is a systemic disease,
the genetic and epigenetic anomalies occurring in the early stage
of carcinogenesis may have already existed in the tumor-adjacent
tissues. These results indicated that RASSF1A gene
methylation may be involved during the occurrence and development
of HCC, as an early event and most likely representing an early
change in HCC. But given that a significant difference was not
observed in the frequency of RASSF1A methylation in HCC and
adjacent liver tissues, RASSF1A was not considered as one of
the candidate auxiliary biomarkers for HCC diagnosis.
In summary, by analysis of the methylation frequency
of 4 TSGs in HCC, tumor-adjacent, and normal liver tissues, the
present study revealed a progressive epigenetic change during the
formation of HCC, which reflected the molecular mechanism of the
multi-step and multi-stage origin of HCC. In addition, the results
demonstrated that the status of RIZ1 and GSTP1 gene
methylation has good specificity for HCC, may better distinguish
between HCC and non-cancerous tissues, and may therefore be used as
novel candidate biomarkers to assist the early screening and
diagnosis of HCC. With further study on TSG methylation and larger
sample sizes in the future, a more thorough insight may be gained
into the effect of abnormal methylation of TSGs on the occurrence
and development of HCC.
Acknowledgements
The authors wish to acknowledge the excellent
technical support of Dr Guang-Hua Luo and Dr Bao-Qiang Wu.
References
|
1
|
Bonasio R, Tu S and Reinberg D: Molecular
signals of epigenetic states. Science. 330:612–616. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herceg Z and Paliwal A: Epigenetic
mechanisms in hepatocellular carcinoma: How environmental factors
influence the epigenome. Mutat Res. 727:55–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Holliday R and Grigg GW: DNA methylation
and mutation. Mutat Res. 285:61–67. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamanaka M, Watanabe M, Yamada Y, Takagi
A, Murata T, Takahashi H, Suzuki H, Ito H, Tsukino H, Katoh T, et
al: Altered methylation of multiple genes in carcinogenesis of the
prostate. Int J Cancer. 106:382–387. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hoque MO, Begum S, Topaloglu O, Jeronimo
C, Mambo E, Westra WH, Califano JA and Sidransky D: Quantitative
detection of promoter hypermethylation of multiple genes in the
tumor, urine and serum DNA of patients with renal cancer. Cancer
Res. 64:5511–5517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guan CN, Chen XM, Lou HQ, Liao XH, Chen BY
and Zhang PW: Clinical significance of axin and β-catenin protein
expression in primary hepatocellular carcinomas. Asina Pac J Cancer
Prev. 13:677–681. 2012. View Article : Google Scholar
|
|
7
|
Zhang YJ, Chen Y, Ahsan H, Lunn RM, Chen
SY, Lee PH, Chen CJ and Santella RM: Silencing of glutathione
S-transferase P1 by promoter hypermethylation and its relationship
to environmental chemical carcinogens in hepatocellular carcinoma.
Cancer Lett. 221:135–143. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tchou JC, Lin X, Freije D, Isaacs WB,
Brooks JD, Rashid A, De Marzo AM, Kanai Y, Hirohashi S and Nelson
WG: GSTP1 CpG island DNA hypermethylation in hepatocellular
carcinomas. Int J Oncol. 16:663–676. 2000.PubMed/NCBI
|
|
9
|
Jin M, Piao Z, Kim NG, Park C, Shin EC,
Park JH, Jung HJ, Kim CG and Kim H: p16 is a major inactivation
target in hepatocellular carcinoma. Cancer. 89:60–68. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhong S, Yeo W, Tang MW, Wong N, Lai PB
and Johnson PJ: Intensive hypermethylation of the CpG island of Ras
association domain family 1A in hepatitis B virus-associated
hepatocellular carcinomas. Clin Cancer Res. 9:3376–3382.
2003.PubMed/NCBI
|
|
11
|
Yamane B and Weber S: Liver-directed
treatment modalities for primary and secondary hepatic tumors. Surg
Clin North Am. 89:97–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Delis SG, Bakoyiannis A, Tassopoulos N,
Athanassiou K, Kelekis D, Madariaga J and Dervenis C: Hepatic
resection for hepatocellular carcinoma exceeding Milan criteria.
Surg Oncol. 19:200–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nakazato H, Suzuki K, Matsui H, et al:
Association of genetic polymorphisms of glutathione-S-transferase
genes (GSTM1, GSTT1 and GSTP1) with familial prostate cancer risk
in a Japanese population. Anticancer Res. 23:2897–2902.
2003.PubMed/NCBI
|
|
14
|
Narimatsu T, Tamori A, Koh N, Kubo S,
Hirohashi K, Yano Y, Arakawa T, Otani S and Nishiguchi S: p16
promoter hypermethylation in human hepatocellular carcinoma with or
without hepatitis virus infection. Intervirology. 47:26–31. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chadwick RB, Jiang GL, Bennington GA, Yuan
B, Johnson CK, Stevens MW, Niemann TH, Peltomaki P, Huang S and de
la Chapelle A: Candidate tumor suppressor RIZ is frequently
involved in colorectal carcinogenesis. Proc Natl Acad Sci USA.
97:2662–2667. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen LB, Xu JY, Yang Z and Wang GB:
Silencing SMYD3 in hepatoma demethylates RIZ1 promoter induces
apoptosis and inhibits cell proliferation and migration. World J
Gastroenterol. 13:5718–5724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang HW, Chan A, Kwong DL, Wei WI, Sham
JS and Yuen AP: Detection of hypermethylated RIZ1 gene in primary
tumor, mouth and throat rinsing fluid, nasopharyngeal swab and
peripheral blood of nasopharyngeal carcinoma patient. Clin Cancer
Res. 9:1033–1038. 2003.PubMed/NCBI
|
|
18
|
Nomoto S, Kinoshita T, Kato K, Otani S,
Kasuya H, Takeda S, Kanazumi N, Sugimoto H and Nakao A:
Hypermethylation of multiple genes as clonal markers in
multicentric hepatocellular carcinoma. Br J Cancer. 97:1260–1265.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Damman R, Li C, Yoon JH, Chin PL, Bates S
and Pfeifer GP: Epigenetic inactivation of RAS association domain
family protein from the lung tumor suppressor locus 3p21.3. Nat
Genet. 25:315–319. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu J, Ni M, Xu J, Zhang H, Gao B, Gu J,
Chen J, Zhang L, Wu M, Zhen S and Zhu J: Methylation profiling of
twenty promoter-CpG islands of genes which may contribute to
hepatocellular carcinogenesis. BMC Cancer. 2:292002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ and
Kang GH: Aberrant CpG island hypermethylation along multistep
hepatocarcinogenesis. Am J Pathol. 163:1371–1378. 2003. View Article : Google Scholar : PubMed/NCBI
|
















