Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
most common entity of the upper aerodigestive tract (1). Carcinoma of this area is the fourth most
common region of cancer incidence and the second most common region
for cancer-related mortality worldwide (2). The intramural heterogeneity may be an
explanation for the worse survival rates recorded in HNSCC. Cancer
stem cells (CSCs) are the most important subpopulation within
carcinoma. CSCs are associated with a worse response to therapy,
metastasis and cancer relapse (3).
Aldehyde dehydrogenase 1 family, member A1 (ALDH1A1) and hyaluronan
receptor cluster of differentiation 44 (CD44) are common markers
for CSCs in HNSCC (4–6).
ALDH1A1 belongs to the ALDH enzyme family and
catalyzes the oxidation of aldehydes to carboxylic acids (7). This enzyme mediates protection from
chemotherapy and reactive oxygen species (ROS) (7,8).
CD44 is an important hyaluronan receptor. CD44 acts
in cell aggregation, proliferation and migration (9). In HNSCC, CD44 is associated with tumor
invasion (10) and the poor survival
of tumor patients (11).
The present study analyzed the expression of ALDH1A1
and CD44 for their relevance in human primary HNSCC. The prognostic
value of ALDH1A1 expression was estimated. Moreover, the utility of
ALDH1A1 and CD44 was evaluated in order to identify CSCs. To define
the CSC population and its activity more precisely, features such
as proliferation (Ki-67) and growth receptor expression [epidermal
growth factor receptor (EGFR)] were included into the analysis.
Materials and methods
Patients
In this study, 48 patients with primary HNSCC (35
male and 13 female) were investigated. Tumor samples were collected
between 1997 and 2008. Experiments were approved by the Ethics
Committee of the Department of Medicine of the Johann Wolfgang
Goethe-University, Frankfurt am Main (276/12).
Sample preparation and analysis
All available samples of each primary tumor were
examined. For immunohistochemistry, formaldehyde-fixed
paraffin-embedded tumor samples, cut to a size of 5 µm, were used.
Antigen retrieval was performed in boiling citrate buffer (pH 6.0;
S1699; Dako, Glostrup, Denmark). ALDH1A1 and CD44 are common
markers for the identification of CSC (4–6),
therefore, these markers were used to identify tumor cell
subpopulations comprised of CSCs. The expression of anti-human
rabbit monoclonal ALDH1A1 (1/100, ab52492; Abcam, Cambridge, UK),
anti-human mouse monoclonal CD44 (1/100, MU310-UC; BioGenex,
Fremont, CA, USA), anti-human mouse monoclonal EGFR (1/50, ab49716;
Abcam) and anti-human rabbit monoclonal Ki-67 (1/200, KI68IC01:
DCS, Hamburg, Germany) were detected. The samples were incubated
with primary antibody for 1 h at room temperature. In the next step
of immunohistochemistry staining procedure, the DCS Detection Line
system staining kits (AD050POL-K, PD000RP and DD006RAP; DCS,
Hamburg, Germany) were used. Staining was developed with DAB
reagent (DC137C100; DCS) and a Fuchsin-Substrate-Chromogen System
(K0625; Dako). Images were captured with a Zeiss Axioplan2 (AxioCam
ICc1 camera; Zeiss, Oberkochen, Germany). The staining intensity of
CD44 and EGFR in the tumor cells was scaled as follows: Strong
(+++), moderate (++), weak (+) and none (−). Weak staining was
described as less intensive and/or discontinuous membrane staining
of the tumor cells.
So far as documented, the clinical history of the
patients and the tumor-node-metastasis (TNM) status (12) of the carcinoma were analyzed.
Statistical analysis
The statistical analysis of data was performed with
BiAS software for Windows (version 10.12; Epsilon-Verlag,
Frankfurt, Germany). The significance between CD44 and EGFR was
analyzed by Yates-Cochram test of trends. The significance of
cancer relapse for the ALDH1A1 expression groups (+/-) was analyzed
by Fisher's exact test. P<0.05 was considered to indicate a
statistically significant difference.
Results
General ALDH1A1 and CD44
expression
The majority of primary HNSCC tumors (45/48; 93.8%)
expressed a minimum of one protein (Table
I). Only three samples (6.3%) expressed neither ALDH1A1 nor
CD44. CD44 expression was detected in 89.6% (43/48) of studied
tumors. CD44 expression intensity was scaled within the tumor cells
(Table II). In 55.8% (24/43) of the
CD44+ tumors expression for the receptor was strong
(+++), in 32.6% (14/43) the CD44 expression was moderate (++) and
in 11.6% (5/43) of samples the CD44 level was weak (+). In 65.1%
(28/43) of the CD44+ tumors this receptor was expressed
in almost all tumor cells. The majority of CD44+ tumors
(38/43, 88.4%) expressed this receptor at a strong to moderate
level (++/+++). Tumor cell nests were solely CD44+ or
CD44-expressing tumor cells were located in the periphery of the
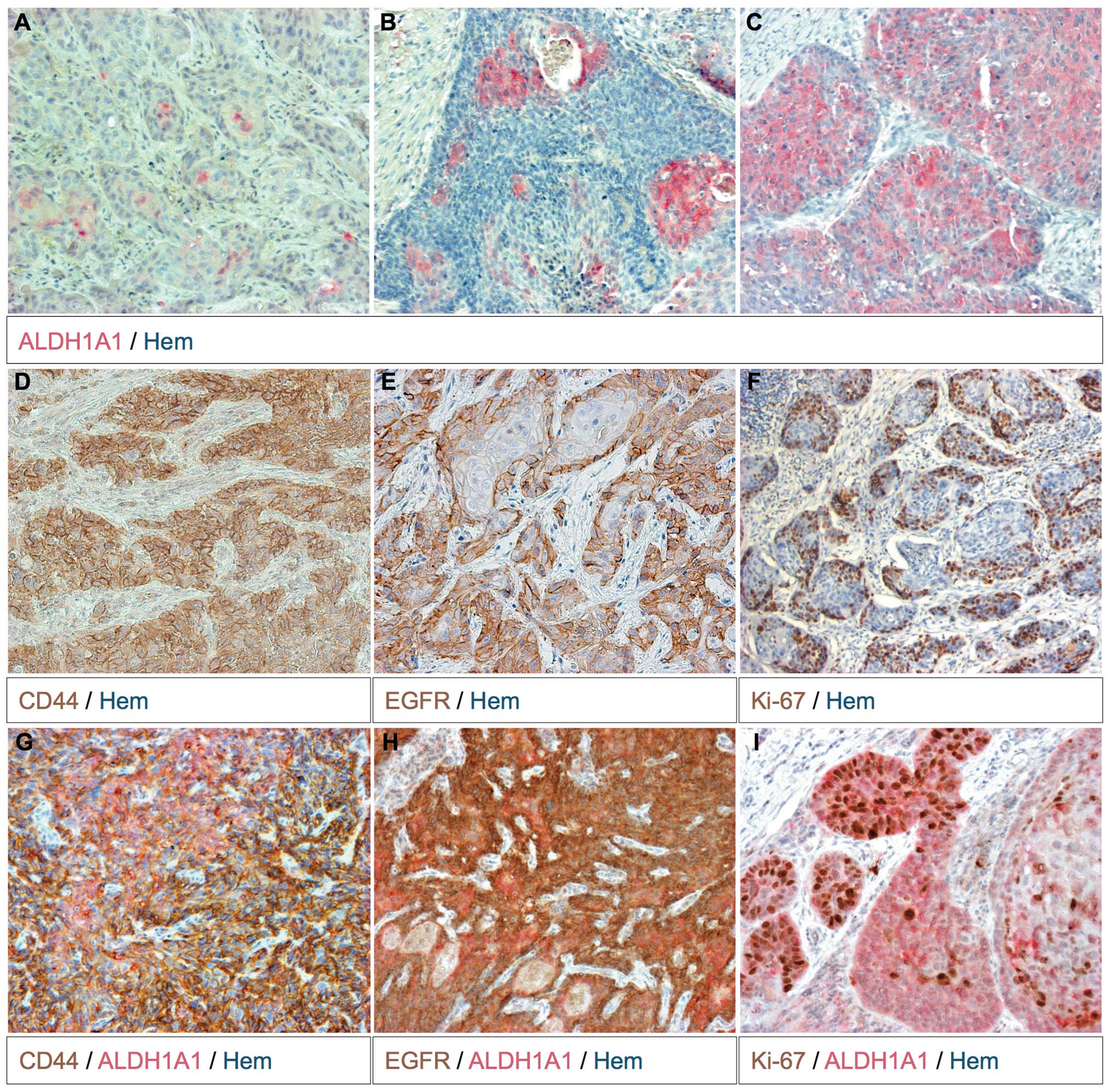
tumor cell nests and close to the stroma (Fig. 1D).
 | Table I.ALDH1A1 and/or CD44 expression in
human primary tumors.a |
Table I.
ALDH1A1 and/or CD44 expression in
human primary tumors.a
|
| ALDH1A1 and/or CD44
expression, n (%) |
|---|
|
|
|
|---|
| Expression |
ALDH1A1+ | CD44
+ |
ALDH1A1+/CD44+ |
ALDH1A1−/CD44− |
|---|
| General
expression | 26 (54.2) | 43 (89.6) | 24 (50.0) | 3 (6.3) |
| Majority
ALDH1A1+ | 8
(30.8) | 8
(18.6) | 8
(33.3) | - |
| Minority
ALDH1A1+ | 18 (69.2) | 16 (37.2) | 16 (66.7) | - |
|
ALDH1A1− | - | 19 (44.2) | - | 3 (100.0) |
| Majority
CD44+ | 13 (50.0) | 28 (65.1) | 11 (45.8) | - |
| Minority
CD44+ | 11 (42.3) | 15 (34.9) | 13 (54.2) | - |
|
CD44− | 2 (7.7) | - | - | 3 (100.0) |
 | Table II.Staining intensity of CD44 and EGFR
in human primary tumors. Linear regression of expression
(P≤0.01). |
Table II.
Staining intensity of CD44 and EGFR
in human primary tumors. Linear regression of expression
(P≤0.01).
|
| Staining intensity
of CD44 and EGFR, n (%) |
|---|
|
|
|
|---|
| Protein | - | + | ++ | +++ |
|---|
| EGFR | 1 (2.1) | 12 (25.0) | 9
(18.8) | 26 (54.2) |
| CD44 | 5 (10.4) | 5
(10.4) | 14 (29.2) | 24 (50.0) |
ALDH1A1 expression was detected in 54.2% (26/48) of
the studied tumor samples (Table I).
In 30.8% (8/26) of the ALDH1A1+ tumors this enzyme was
expressed in the majority of tumor cells (Fig. 1C). In the remaining
ALDH1A1+ tumors (18/26, 69.2%) ALDH1A1-expressing cells
were found to be singular, in groups or throughout the entirely
cell nest (Fig. 1A and B). More often
ALDH1A1+ tumor cell groups were located more centrally
within the cell nests (Fig. 1A and
B).
Half (24/48) of the analyzed tumors expressed
ALDH1A1 and CD44 (Table I). In these
tumors the expression pattern of ALDH1A1 and CD44 intersected, but
did not completely overlap (Fig. 1G).
With the exception of one tumor (7/8, 87.5%), the human papilloma
virus (HPV)+ tumors expressed ALDH1A1 and CD44. The
remaining HPV+ tumor showed CD44 staining (Table III).
 | Table III.Evaluation of available patient data
for ALDH1A1 expression and HPV status.a |
Table III.
Evaluation of available patient data
for ALDH1A1 expression and HPV status.a
|
| Evaluation of
patient data, n (%) |
|---|
|
|
|
|---|
| Factor |
ALDH1A1+ |
ALDH1A1+/HPV− |
ALDH1A1− |
ALDH1A1−/HPV− |
|---|
| Total patients | 26 (100.0) | 19 (100.0) | 22 (100.0) | 21 (100.0) |
| Gender |
|
|
|
|
|
Male | 20 (76.9) | 14 (73.7) | 15 (68.2) | 15 (71.4) |
|
Female | 6
(23.1) | 5
(26.3) | 7
(31.8) | 7
(33.3) |
| Cancer relapse |
|
|
|
|
|
Positive | 6
(23.1) | 6
(31.6) | 12 (54.5) | 12 (57.1) |
|
Negative | 20 (76.9) | 13 (68.4) | 10 (45.5) | 9
(42.9) |
| Age, years |
|
|
|
|
|
<60 | 10 (38.5) | 7
(36.8) | 13 (59.1) | 12 (57.1) |
|
60–70 | 9
(34.6) | 9
(47.4) | 7
(31.8) | 7
(33.3) |
|
>70 | 7
(26.9) | 3
(15.8) | 2
(9.1) | 2
(9.5) |
| T stage | 25 (100.0) | 18 (100.0) | 18 (100.0) | 17 (100.0) |
| T1 | 0
(0.0) | 0
(0.0) | 4
(22.2) | 4
(23.5) |
| T2 | 12 (48.0) | 9
(50.0) | 7
(38.9) | 7
(41.2) |
| T3 | 9
(36.0) | 5
(27.8) | 5
(27.8) | 4
(23.5) |
| T4 | 4
(16.0) | 4
(22.2) | 2
(11.1) | 2
(11.8) |
| N stage | 24 (100.0) | 17 (100.0) | 18 (100.0) | 17 (100.0) |
| N0 | 8
(33.3) | 4
(23.5) | 6
(33.3) | 6
(35.3) |
|
N1-N3 | 16 (66.7) | 13 (76.5) | 12 (66.7) | 11 (64.7) |
EGFR and Ki67 status of
ALDH1A1+ or CD44+ tumor cells
With the exception of one tumor, the examined HNSCC
tumors expressed EGFR (47/48, 97.9%) (Table II). In most tumors the majority of
tumor cells were EGFR+. The staining intensity of EGFR
was scaled as strong to moderate (++/+++) in 74.5% (35/47) of
EGFR+ tumors. The staining patterns of CD44 and EGFR
were mostly overlapping. The position of the EGFR+ and
CD44+ tumor cells was generally identical. However,
EGFR+ tumor cells were located more marginally within
the tumor cell nests (Fig. 1E). The
association between EGFR and CD44 staining intensity could be
described as an linear regression (P≤0.01). In the majority of
tumors, ALDH1A1+ tumor cells were not necessarily
CD44+ (Fig. 1G). EGFR
staining gave similar results (Fig.
1H). The amount of Ki-67+ tumor cells also appeared
to be independent of ALDH1A1 expression (Fig. 1I). Closer to the stroma the
proliferation rate was higher (Fig.
1F).
Analysis of ALDH1A1+ and
ALDH1A1− patients
The patient collective was divided into
ALDH1A1+ and ALDH1A1− primary tumor samples.
Notably, the majority of confirmed tonsillar tumors (11/16; 68.8%)
were ALDH1A1+. Due to the low number of CD44−
tumors (5/48; 10.4%), an analysis of CD44+ and
CD44− samples was not meaningful.
The appearance of metastasis and relapse was
analyzed (Table III). In the
majority of cases, cancer relapse was observed up to 2 years after
primary tumor detection. Cancer relapse occurred in 23.1% (6/26) of
patients with ALDH1A1+ primary tumors. The number of
relapses was significantly higher (P≤0.05) for patients with
ALDH1A1− primary tumors (12/22; 54.5%). No notable
differences were found between the TNM stage of ALDH1A1+
and ALDH1A1− tumors. However, one explanation for the
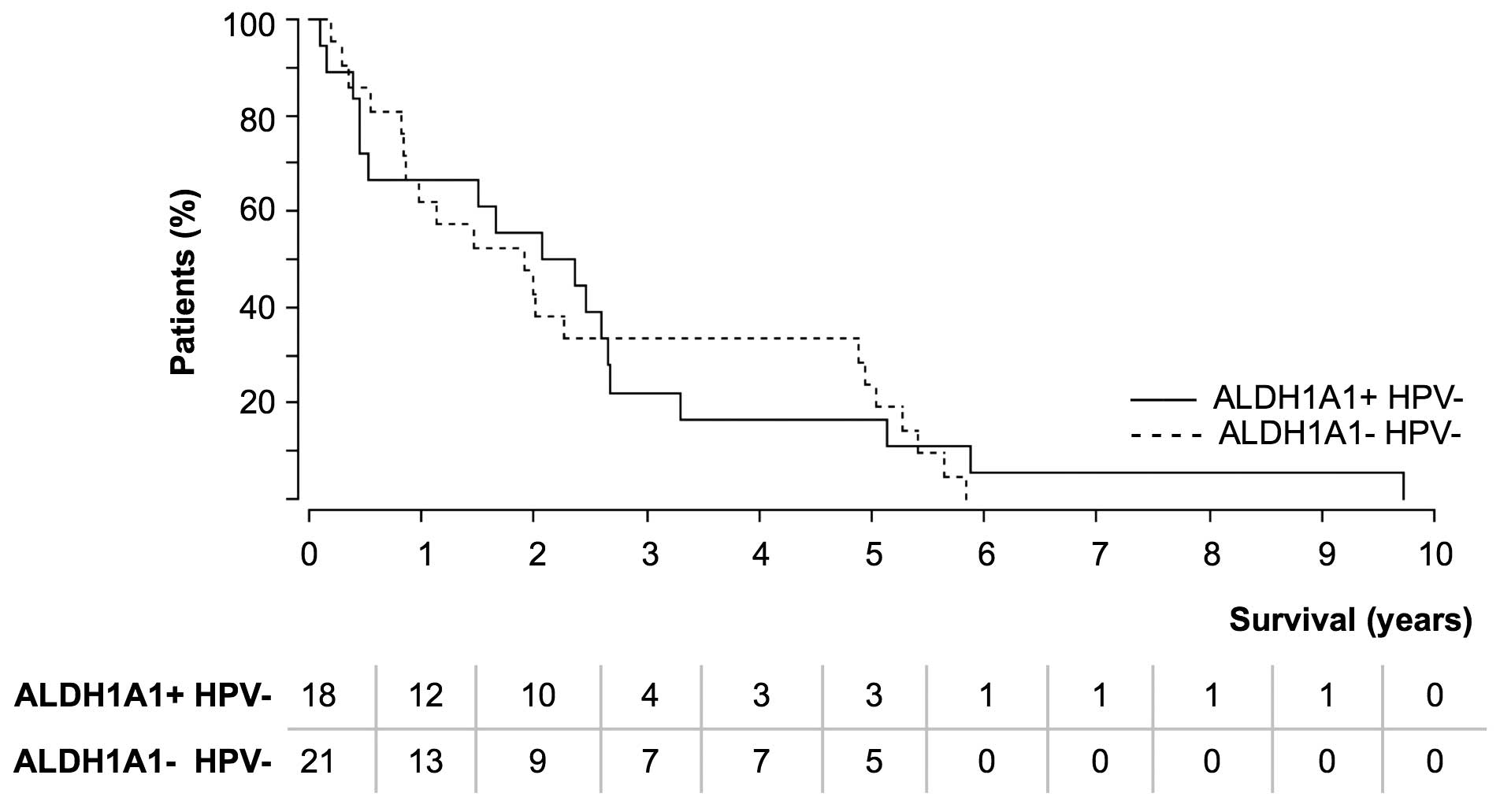
increased number of relapses resulted in the comparison of overall
survival (Fig. 2). As HPV is a
prognostic factor for longer survival (13), HPV+ cases were
excluded.
The majority of patients in the two groups succumbed
within the first 3 years of tumor appearance. Within the
ALDH1A1+ (HPV−) tumor group, fewer patients
(4/18; 22.2%) survived longer than 3 years compared with the
ALDH1A1− (HPV−) tumor group (7/21; 33.3%).
This issue could not be explained by a younger age of the
ALDH1A1− tumor patients, as the analysis of different
age groups (<60, 60–70 and >70 years) gave comparable results
(data not shown). Survival was not linked to the percentage of
ALDH1A1+ cells within the tumor.
Discussion
ALDH1A1 and CD44 function in the carcinogenesis and
tumor progression of HNSCC. ALDH1A1 is not expressed in the normal
oral mucosa (14,15). However, other ALDH isoenzymes, such as
ALDH1A3 and ALDH3A1, are found (14).
The expression of the retinoic acid receptors in the oral mucosa
underlines the function of ALDH isoenzymes in the normal mucosa
(16). However, ALDH1A1 expression is
increased in dysplasia and HNSCC (6).
In the present study, it was found that 54.2% of primary HNSCC
tumors were ALDH1A+. In 30.8% of the ALDH1A1+
tumors, ALDH1A1 was expressed in the majority of tumor cells. This
observations confirm a more general function of ALDH1A1 in the
carcinogenesis of HNSCC. One explanation is that ALDH1A1 is
stress-induced. The ALDH1A1 promoter has binding sites for the AP-1
(17) and Oct-1 (18) transcription factors. Oct-1 is
stabilized and translocated into the nucleus due to radiation, ROS
(19) and HPV16 infection (20). The low expression of ALDH1A1 has also
been recognized in the normal tonsillar squamous epithelium
(21). Additionally, ALDH1A1 protects
from oxidative stress-induced reactive aldehydes (8). In the present study, ALDH1A1+
tumor cells were mostly located in the middle of tumor cell nests
and assumed hypoxic tumor sites. Furthermore, ALDH1A1 appeared to
be a prognostic factor for worse survival. This was in agreement
with the results of a study by Qian et al (22). The reduced relapse rates for patients
with ALDH1A1+ tumors may arise from worse survival
rates. Additionally, the majority of HPV-associated carcinomas are
ALDH1A+, and HPV-induced HNSCC exhibits a better
prognosis (13). These observations
possibly verify the role of ALDH1A1 in tumor progression.
CD44 is expressed in the basal layer of the normal
oral mucosa (21,23–25). As a
consequence, CD44 was overexpressed in many of the examined tumors
in the present study. CD44 was located entirely throughout the cell
nest or peripheral to the stroma. CD44 can be linked to the
invasion of HNSCC (10). However, the
present results indicate that CD44 has a more general function in
carcinogenesis. CD44 was often coexpressed with EGFR. Additionally
the expression intensities of CD44 and EGFR were correlated
significantly throughout the tumor. It is known that CD44 can
interact with tyrosine kinase receptors, such as EGFR (26). Interaction of these surface receptors
can cause proliferation, chemotherapy resistance and invasion in
HNSCC cell lines (27,28). EGFR also increases CD44 mRNA
expression (29). Additionally,
proliferating tumor cells (Ki-67) are often CD44+.
Consequently, CD44 also has a significant role in HNSCC tumor
progression.
ALDH1A1 and CD44 are common CSC markers in HNSCC
(4–6).
The present study examined the ability of the two proteins to
identify CSCs. Corresponding to the hierarchical tumor model, CSCs
exist in every tumor (30,31). In the present study, the ALDH1A1 and
CD44 proteins were expressed in the majority of tumors and also in
HPV+ carcinoma. Consequently, ALDH1A1 and CD44 could
also be expressed in the CSCs of these tumors. As a CSC marker,
CD44 was more often expressed in the tumors than ALDH1A1. However,
a lot of tumors expressed CD44 in almost all tumor cells. To
identify CSCs, the markers must isolate CSCs from the tumor bulk.
ALDH1A1 was a better marker to define a subpopulation of tumor
cells. Finally, the two markers were not sufficient to isolate the
CSCs from the bulk of tumor cells. Further CSC markers should be
used to define and isolate the CSC population.
In addition, ALDH1A1 und CD44 expression did not
completely overlap. In the majority of tumors
ALDH1A1+/CD44+,
ALDH1A1+/CD44− and
ALDH1A1−/CD44+ populations were observed.
This observation may indicate that different CSC populations could
exist within one tumor. For HNSCC, this theory of different CSC
phenotypes was first suggested by Biddle et al (32). This hypothesis has also been discussed
for breast (33,34) and colorectal (30) cancer. Different phenotypes would make
it more difficult to determine the CSC population.
In conclusion, CD44 and ALDH1A1 appear to be
important factors in carcinogenesis and tumor progression. ALDH1A1
was shown to be a possible prognostic marker for worse survival,
while ALDH1A1 and CD44 may be expressed in the CSCs of most
examined tumors. However, these markers are not sufficient to
precisely isolate the CSC subpopulation from the tumor bulk.
Acknowledgements
This study was funded by a young investigator grant
of UCT Frankfurt/Main and the Heinrich and Erna Schaufler-Stiftung
(grant no. SKHT-HNO-01/13).
References
|
1
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012, cancer incidence and mortality worldwide:
IARC cancerbase No. 11 [Internet]. Lyon, France: IARC; 1.0.
2013
|
|
3
|
Wu MJ, Jan CI, Tsay YG, Yu YH, Huang CY,
Lin SC, Liu CJ, Chen YS, Lo JF and Yu CC: Elimination of head and
neck cancer initiating cells through targeting glucose regulated
protein78 signaling. Mol Cancer. 9:2832010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen YC, Chen YW, Hsu HS, Tseng LM, Huang
PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al: Aldehyde
dehydrogenase 1 is a putative marker for cancer stem cells in head
and neck squamous cancer. Biochem Biophys Res Commun. 385:307–313.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clay MR, Tabor M, Owen JH, Carey TE,
Bradford CR, Wolf GT, Wicha MS and Prince ME: Single-marker
identification of head and neck squamous cell carcinoma cancer stem
cells with aldehyde dehydrogenase. Head Neck. 32:1195–1201. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visus C, Ito D, Amoscato A,
MaciejewskaFranczak M, Abdelsalem A, Dhir R, Shin DM, Donnenberg
VS, Whiteside TL and DeLeo AB: Identification of human aldehyde
dehydrogenase 1 family member A1 as a novel CD8+ T-cell-defined
tumor antigen in squamous cell carcinoma of the head and neck.
Cancer Res. 67:10538–10545. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma I and Allan AL: The role of human
aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell
Rev. 7:292–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh S, Brocker C, Koppaka V, Chen Y,
Jackson BC, Matsumoto A, Thompson DC and Vasiliou V: Aldehyde
dehydrogenases in cellular responses to oxidative/electrophilic
stress. Free Radic Biol Med. 56:89–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Turley EA, Noble PW and Bourguignon LY:
Signaling properties of hyaluronan receptors. J Biol Chem.
277:4589–4592. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang SJ and Bourguignon LY: Role of
hyaluronan-mediated CD44 signaling in head and neck squamous cell
carcinoma progression and chemoresistance. Am J Pathol.
178:956–963. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kokko LL, Hurme S, Maula SM, Alanen K,
Grénman R, Kinnunen I and Ventelä S: Significance of site-specific
prognosis of cancer stem cell marker CD44 in head and neck
squamous-cell carcinoma. Oral Oncol. 47:510–516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gillespie MB, Rubinchik S, Hoel B and
Sutkowski N: Human papillomavirus and oropharyngeal cancer: What
you need to know in 2009. Curr Treat Options Oncol. 10:296–307.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hedberg JJ, Grafström RC, Vondracek M,
Sarang Z, Wärngård L and Höög JO: Micro-array chip analysis of
carbonyl-metabolising enzymes in normal, immortalised and malignant
human oral keratinocytes. Cell Mol Life Sci. 58:1719–1726. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kato H, Izumi K, Saito T, Ohnuki H, Terada
M, Kawano Y, NozawaInoue K, Saito C and Maeda T: Distinct
expression patterns and roles of aldehyde dehydrogenases in normal
oral mucosa keratinocytes: Differential inhibitory effects of a
pharmacological inhibitor and RNAi-mediated knockdown on cellular
phenotype and epithelial morphology. Histochem Cell Biol.
139:847–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sherman JA and Partridge M: Expression of
retinoic acid receptors in normal, dysplastic and malignant oral
epithelia. Br J Oral Maxillofac Surg. 35:260–266. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Makia NL, Amunom I, Falkner KC, Conklin
DJ, Surapureddi S, Goldstein JA and Prough RA: Activator protein-1
regulation of murine aldehyde dehydrogenase 1a1. Mol Pharmacol.
82:601–613. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maddox J, Shakya A, South S, Shelton D,
Andersen JN, Chidester S, Kang J, Gligorich KM, Jones DA, Spangrude
GJ, et al: Transcription factor Oct1 is a somatic and cancer stem
cell determinant. PLoS Genet. 8:e10030482012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang P and Jin T: Oct-1 functions as a
sensor for metabolic and stress signals. Islets. 2:46–48. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong HK and Ziff EB: The human
papillomavirus type 16 E7 protein complements adenovirus type 5 E1A
amino-terminus-dependent transactivation of adenovirus type 5 early
genes and increases ATF and Oct-1 DNA binding activity. J Virol.
70:332–340. 1996.PubMed/NCBI
|
|
21
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based Human Protein Atlas. Nat
Biotechnol. 28:1248–1250. 2010.Available from. http://www.proteinatlas.org View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qian X, Wagner S, Ma C, Coordes A, Gekeler
J, Klussmann JP, Hummel M, Kaufmann AM and Albers AE: Prognostic
significance of ALDH1A1-positive cancer stem cells in patients with
locally advanced, metastasized head and neck squamous cell
carcinoma. J Cancer Res Clin Oncol. 140:1151–1158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mack B and Gires O: CD44 s and CD44v6
expression in head and neck epithelia. PLoS One. 3:e33602008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sterz CM, Kulle C, Dakic B, Makarova G,
Böttcher MC, Bette M, Werner JA and Mandic R: A basal-cell-like
compartment in head and neck squamous cell carcinomas represents
the invasive front of the tumor and is expressing MMP-9. Oral
Oncol. 46:116–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Richard V and Pillai MR: The stem cell
code in oral epithelial tumorigenesis: ‘the cancer stem cell shift
hypothesis’. Biochim Biophys Acta. 1806:146–162. 2010.PubMed/NCBI
|
|
26
|
Williams K, Motiani K, Giridhar PV and
Kasper S: CD44 integrates signaling in normal stem cell, cancer
stem cell and (pre)metastatic niches. Exp Biol Med. 238:324–338.
2013. View Article : Google Scholar
|
|
27
|
Wang SJ and Bourguignon LY: Hyaluronan and
the interaction between CD44 and epidermal growth factor receptor
in oncogenic signaling and chemotherapy resistance in head and neck
cancer. Arch Otolaryngol Head Neck Surg. 132:771–778. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bourguignon LY, Gilad E, Brightman A,
Diedrich F and Singleton P: Hyaluronan-CD44 interaction with
leukemia-associated RhoGEF and epidermal growth factor receptor
promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+
signaling and cytoskeleton modification in head and neck squamous
cell carcinoma cells. J Biol Chem. 281:14026–14040. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abhold EL, Kiang A, Rahimy E, Kuo SZ,
WangRodriguez J, Lopez JP, Blair KJ, Yu MA, Haas M, Brumund KT, et
al: EGFR kinase promotes acquisition of stem cell-like properties:
a potential therapeutic target in head and neck squamous cell
carcinoma stem cells. PLoS One. 7:e324592012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brabletz T, Jung A, Spaderna S, Hlubek F
and Kirchner T: Opinion: Migrating cancer stem cells-an integrated
concept of malignant tumour progression. Nat Rev Cancer. 5:744–749.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Biddle A, Liang X, Gammon L, Fazil B,
Harper LJ, Emich H, Costea DE and Mackenzie IC: Cancer stem cells
in squamous cell carcinoma switch between two distinct phenotypes
that are preferentially migratory or proliferative. Cancer Res.
71:5317–5326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu S, Clouthier SG and Wicha MS: Role of
microRNAs in the regulation of breast cancer stem cells. J Mammary
Gland Biol Neoplasia. 17:15–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
French R and Clarkson R: The complex
nature of breast cancer stem-like cells: Heterogeneity and
plasticity. J Stem Cell Res Ther. S7:0092012.
|