Introduction
Esophageal squamous cell carcinoma (ESCC) is a
malignant disease with uneven geographical distribution worldwide.
Notably, the age of patients at the time of diagnosis is reducing
(1). In East Asia (China, Japan and
South Korea), ESCC has high incidence and mortality rates, and
accounts for 90% of all cases of esophageal carcinoma (2). Despite advances in combined modality
therapies for ESCC, including surgery, chemotherapy and
radiotherapy, the prognosis for patients remains poor, and the
5-year overall survival rate is 20–30% (3). Although a number of genetic and
epigenetic alterations associated with ESCC have been described
(for example, p53, Bcl-2, epidermal growth factor receptor, RUNX3
and esophageal cancer related gene 1) (4–6), its
oncogenesis and pathogenesis remain largely unknown. Optimal and
commonly-used molecular markers to facilitate the comprehensive
management of patients, including early diagnosis and prognostic
evaluation, have not been proposed thus far. The prognosis of ESCC
patients is tumor-node-metastasis (TNM) stage-specific, but the TNM
stage is not sufficiently sensitive to evaluate the prognosis of
patients (7). Therefore, the
discovery of biomarkers for therapeutic development and prognostic
prediction is required.
ESCC is characterized by genome instability through
severe DNA damage caused by several factors, including the
consumption of food or beverages at high temperatures, tobacco
smoking, poor nutrition, infection and heredity (8). Ku80, one of the subunits of the
Ku80/Ku70 heterodimer, is a central element in the nonhomologous
end joining (NHEJ) DNA repair pathway. Double-strand DNA breaks
(DSB) activate the catalytic subunit of the DNA-dependent protein
kinase and trigger NHEJ repair activities (9). It has been demonstrated that the
silencing of Ku80 proteins prevents DSB repair and telomere
maintenance, and results in cell cycle arrest, apoptosis,
chemosensitivity and radiosensitivity (10,11).
Previous studies have demonstrated that Ku80 is associated with
pathological processes in certain malignant tumors, and have
provided valuable information regarding the expression levels of
Ku80 and its pathological role in ESCC (11–14). Yang
et al (11) reported that Ku80
participates in tumorigenesis, radioresistance and chemoresistance
in esophageal cancer cells. Tonotsuka et al (14) observed that Ku80 protein was expressed
in the nuclei of basal cell layers and luminal cell layers in
esophageal cancer tissues. However, the protein expression pattern
of Ku80 and its clinicopathological significance in ESCC is not
well established. In the present study, the expression levels of
Ku80 in ESCC tissues were analyzed, and the association of Ku80
expression with the clinicopathological features and prognosis of
patients affected with ESCC were further investigated.
Materials and methods
Ethics statement
The current study protocol was reviewed and approved
by the Research Ethics Committee of the Provincial Hospital
Affiliated to Shandong University (Jinan, China) (2003–063). All
participants provided written informed consent for the detection of
Ku80 in the tissue-derived samples, subsequent data analysis and
publication of the results.
Patients and samples
From January until May 2003, 119 patients with ESCC
(41 females and 78 males; mean age, 57.8±12.3 years) were screened
in the Provincial Hospital Affiliated to Shandong University. These
patients were diagnosed with ESCC by histopathological detection at
the Pathology Department of the hospital, precluding esophageal
leiomyoma or other benign disease and malignant tumors originated
from organs other than the esophagus. Eligibility was granted if
primary diagnosis occurred ≤6 months prior to study enrollment and
patients had not received chemotherapy, radiotherapy or biotherapy
prior to sample collection. Full medical examinations were
recorded, and the clinicopathological characteristics of the
patients were analyzed. All the ESCC patients included in the
present study were restaged according to the 2009 International
Union Against Cancer TNM staging guidelines for esophageal cancer
(15). The histopathological
evaluation of the samples was performed according to the criteria
proposed by the World Health Organization (16).
In the control group, 109 volunteers (35 females and
74 males; mean age, 56.6±13.2 years) from the Provincial Hospital
Affiliated to Shandong University were screened as normal subjects
without malignant disease. Their medical records indicated the
absence of drug, tobacco and alcohol abuse. All individuals were
Han Chinese without consanguineous relationships. Detailed
questionnaires were completed by the subjects participating in the
present study. The questionnaires collected information about the
individuals, including medical and family history, use of
over-the-counter medications and exposure to dietary carcinogens.
The detailed questionnaires were used to measure average dietary
intake 1 year prior to the date of selection for the current study.
No significant differences were observed between the 119 ESCC
patients and the 109 healthy volunteers (regarding age, gender,
medical and family history, smoking status, exposure to dietary
carcinogens and dietary habit). All patients fasted ≥12 h and had
not smoked for 6 months prior to the collection of tissue
samples.
In the ESCC group, 119 pairs of samples were
collected from the 119 patients by gastroscopy. Each pair of
samples consisted of ESCC tissue and corresponding healthy mucosa
(CHEM).
The corresponding healthy esophageal mucosa was
harvested from a position >5 cm in distance from the margin of
the ESCC. A total of 109 normal esophageal mucosa (NEM) samples
from the control group were harvested via gastroscopy. The
macroscopic examination of the healthy mucosa revealed no signs of
deterioration and necrosis. Light microscope examination
demonstrated that the healthy mucosal tissues were free of tumor
and any detectable concurrent disease, including esophagitis and
dysplasia. Following encapsulation in tin foil wrapper, the tissue
samples were rinsed in cold NaCl (0.9%), and immediately stored at
−80°C until further use.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from esophageal mucosal
tissues using TRIzol (Invitrogen Life Technologies, Carlsbad, CA,
USA), according to the manufacturer's instructions. The RT system
included 2 µg RNA, oligo-dT15 primer and M-MLV reverse
transcriptase (Takara Bio, Inc., Shiga, Japan) in a volume of 20 µl
diethylpyrocarbonate-treated water (Takara Shuzo Co., Ltd., Kyoto,
Japan). Gene-specific primer sequences (Dingguo Changsheng
Biotechnology Co., Ltd., Beijing, China) were as follows: Ku80, F
5′-ACG ATT TGG TAC AGA TGG CAC T-3′ and R 5′-GCT CCT TGA AGA CGC
ACA GTT T-3′ (product, 497 bp); GAPDH, F 5′-GAA GGT GAA GGT CGG AGT
C-3′ and R 5′-GAA GAT GGT GAT GGG ATT TC-3′ (product, 300 bp).
RT-qPCR was performed in a LightCycler 480 (Roche Diagnostics,
Nutley, NJ, USA) under the following conditions: 1 cycle for 3 min
at 94°C followed by 30 cycles of 30 sec at 94°C, 30 sec at 57°C and
60 sec at 72°C. The reactions were terminated at 4°C, following
5-min elongation at 72°C. The expression levels of GAPDH were used
as an internal control for RNA quantity and quality. The relative
quantity of mRNA (RQ) was calculated as the calibrator-normalized
ratio using the LightCycler 480 software, version 1.5 (Roche
Diagnostics), applying the following formula: RQ =
2−ΔΔCt, where ΔΔCt = (Cttargeted gene -
CtGAPDH)targeted sample - (Cttargeted gene -
CtGAPDH)calibration sample. All the experiments were
repeated ≥3 times, and included controls without cDNA, primers or
thermophilic polymerase. The specificity of the assay was
determined by PCR, using all the primer pairs on each cloned
template cDNA to exclude cross-reactivity.
Immunohistochemistry (IHC)
IHC staining for Ku80 was performed on 4-µm tissue
sample sections using an UltraVision Quanto detection system
(Thermo Fisher Scientific, Inc., Fremont, CA, USA) following the
manufacturer's instructions. Briefly, the sections were incubated
with antigen retrieval solution (pH 7.0; Beijing Solarbio Science
& Technology Co., Ltd., Beijing, China) for 10 min at 95°C
followed by incubation with UltraVision hydrogen peroxide block
(Thermo Fisher Scientific, Inc.) for 10 min to block any endogenous
peroxidase activity. Next, the sections were incubated with
UltraVision protein block (Thermo Fisher Scientific, Inc.) for 5
min to reduce nonspecific background staining. Following overnight
incubation at 4°C with a rabbit monoclonal antihuman Ku80 primary
antibody (1:500; cat. no. ab80592; Abcam, Cambridge, UK), the
specimens were washed with phosphate-buffered saline (PBS), and
incubated with the primary antibody amplifier Quanto (Thermo Fisher
Scientific, Inc.) for 10 min, followed by 10-min incubation with
horseradish peroxidase polymer Quanto (Thermo Fisher Scientific,
Inc.). Next, the sections were washed with PBS and deionized water,
incubated with 3,3-diaminobenzidine (Dingguo Changsheng
Biotechnology Co., Ltd.) and counterstained with hematoxylin
(Beyotime Institute of Biotechnology, Nantong, China). The
specificities of the primary antibodies had been previously
confirmed, and their use as positive controls in HeLa cells has
also been validated, since previous studies have demonstrated that
Ku80 is overexpressed in these cells (17). The control sections were incubated
with PBS instead of the primary antibodies and used as negative
controls.
The immunohistochemical scoring of Ku80 was
performed using a semiquantitative system as previously reported
(18). The specimens were examined
under a light microscope. In 5 randomly selected fields per
section, the number of immunoreactive cells/100 cells was assessed
and quantified as a percentage. Next, the average percentage of the
5 fields was used to assess the proportion score in a 6-category
grading system (0, negative; 1, 1–10%; 2, 11–25%; 3, 26–50%; 4,
51–75%; and 5, >75%). The intensity score for staining was
estimated using a 4-category grading system (0, negative; 1, weak;
2, moderate; and 3, strong). The IHC score was defined as the
proportion score × the intensity score. The scientists were blinded
to the patients data, and scored the samples independently, then
reached agreement by repeated analysis and discussion.
Receiver operating characteristics
(ROC) curve
The cut-off scores for Ku80 mRNA and protein
overexpression levels were screened based on the ROC curve. The raw
data corresponding to Ku80 mRNA and protein expression levels in
the ESCC and the control groups were analyzed by the MedCalc
statistical software package, version 13.0.2.0 (MedCalc Software
bvba, Ostend, Belgium). The score closest to the point with maximum
sensitivity and specificity was selected as the cut-off score
leading to the largest number of patients correctly classified with
or without overexpression of Ku80 mRNA and protein levels.
Statistical analysis
All statistical analyses were performed using SPSS
software, version 17.0 (SPSS Inc., Chicago, IL, USA). Statistical
comparisons between the Ku80 mRNA expression levels in the
different groups were performed by analysis of variance (ANOVA).
The Mann-Whitney U test was used to determine the differences in
Ku80 protein expression levels across the groups. Associations
between categorical variables were analyzed using the χ2
test. The survival time was calculated from the date of the
diagnosis until mortality or the end of the study. The survival
curves were calculated by the Kaplan-Meier method. Univariate
log-rank test and Cox regression model analysis were performed to
identify prognostic factors. A 2-tailed P<0.05 was considered to
indicate a statistically significant difference.
Results
High expression levels of Ku80 mRNA in
ESCC tissues
The mRNA expression levels of Ku80 were examined in
119 pairs of samples from the ESCC group and in 109 samples from
the control group. The relative mRNA expression levels of Ku80 in
ESCC tissue, CHEM and NEM were 5.348±1.480, 3.327±1.106 and
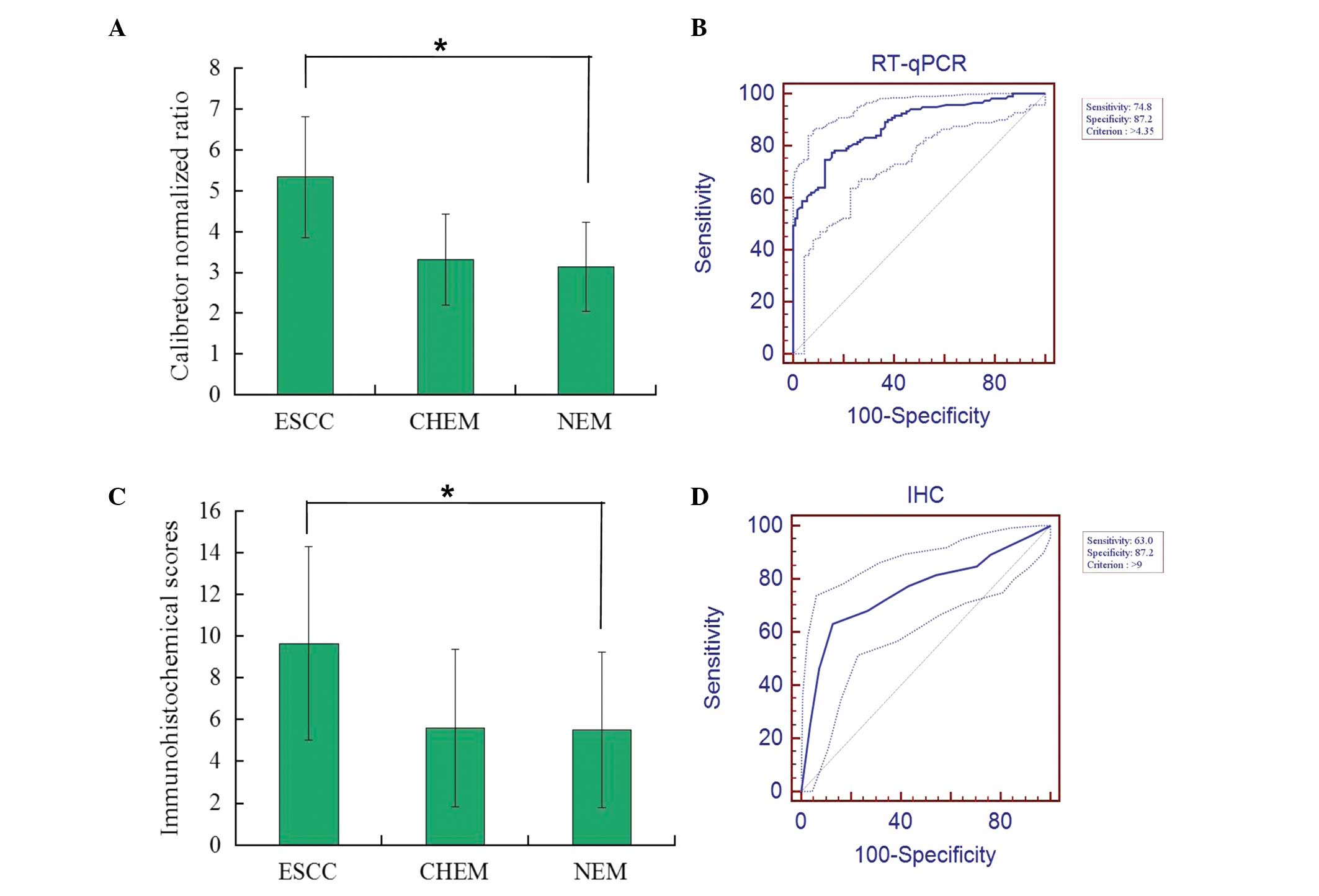
3.149±1.092, respectively (Fig. 1A).
The mRNA levels of Ku80 in ESCC were significantly higher than in
CHEM and NEM (ANOVA; P<0.001). However, no clear difference in
the mRNA levels of Ku80 between CHEM and NEM was observed
(P=0.866). According to the ROC curve (Fig. 1B), the threshold value of 4.35 was the
closest to the point with maximum sensitivity and specificity of
74.8 and 87.2%, respectively; thus, it was selected as the cut-off
value. The area under the ROC curve (AUC) was 0.878 [95% CI,
0.829–0.918]. Therefore, samples with a calibrator-normalized ratio
>4.35 were identified as high expression levels of Ku80 mRNA,
whereas the remaining samples were considered to have low levels.
Consequently, patients were divided into 2 groups; high (n=84,
70.6%) and low (n=35, 29.4%) mRNA expression groups. However, in
the control group there were 20 (18.3%) cases of high expression of
Ku80 mRNA and 89 (81.7%) cases of low expression. Overall, the
frequency of high Ku80 mRNA expression levels was significantly
increased in ESCC compared with NEM (χ2 test;
P<0.001) (data not shown).
High expression levels of Ku80 protein
in ESCC tissues
The IHC scores in ESCC tissue, CHEM and NEM were
9.656±4.633, 5.608±3.759 and 5.532±3.741, respectively (Fig. 1C). The IHC scores of Ku80 in ESCC was
significantly higher than those in CHEM and NEM (Mann-Whitney U
test; P<0.001). However, there was no clear difference in IHC
scores between CHEM and NEM (P=0.268). According to the ROC curve
analysis (Fig. 1D), the cut-off score
was 9, with maximum sensitivity of 63.0% and specificity of 87.2%.
The AUC was 0.756 (95% CI; 0.695–0.811). Consequently, the patients
were divided into 2 groups; high (n=75, 63.0%) and low (n=44,
37.0%) protein expression groups. However, there were 18 (15.1%)
cases of high expression of Ku80 mRNA and 101 (84.9%) cases of low
expression in the control group. The difference in frequency of
high protein expression levels between the ESCC tissues and NEM was
observed to be statistically significant (χ2 test;
P<0.001). Spearman bivariate correlation indicated a positive
correlation between the mRNA and the protein expression levels of
Ku80 (r=0.923; P<0.001) (data not shown). Visual analysis of the
IHC staining was performed, and positive expression of Ku80 protein
was revealed as yellow or brownish yellow stain in the nuclei of
tumor cells. Strong positive Ku80 staining was observed in the
cancer cell nuclei, whereas negative or weak staining was observed
in CHEM and NEM (Fig. 2).
Clinicopathological characteristics of
ESCC patients and Ku80 expression levels
The clinicopathological features (age, gender, tumor
site, tumor size, differentiation degree, local invasion, lymph
node metastasis and TNM stage) of the 119 patients with ESCC in the
current study are summarized in Table
I. χ2 analysis indicated that the Ku80 mRNA
expression levels positively correlated with differentiation degree
(P=0.016), local invasion (P=0.016), lymph node metastasis
(P=0.002) and TNM stage (P=0.001), but not with age (P=0.840),
gender (P=0.980), tumor location (P=0.351) or tumor size (P=0.407)
(Table I). In agreement with the mRNA
expression pattern, high expression levels of Ku80 protein
presented significant correlations with differentiation degree
(P=0.001), local invasion (P=0.004), lymph node metastasis
(P=0.007) and TNM stage (P=0.002), but no correlation was observed
between the protein expression levels and age (P=0.849), gender
(P=0.842), tumor location (P=0.906) or tumor size (P=0.113).
 | Table I.Associations between Ku80 expression
levels and multiple clinicopathological parameters in ESCC. |
Table I.
Associations between Ku80 expression
levels and multiple clinicopathological parameters in ESCC.
|
|
| Ku80 mRNA levels | Ku80 protein
levels |
|---|
|
|
|
|
|
|---|
| Characteristics | Cases (119) | Low (35) | High (84) | P-value | Low (44) | High (75) | P-value |
|---|
| Age (years) |
|
|
| 0.840 |
|
| 0.849 |
| ≥60 | 54 | 15 | 39 |
| 19 | 35 |
|
|
<60 | 65 | 20 | 45 |
| 25 | 40 |
|
| Gender |
|
|
| 0.980 |
|
| 0.842 |
| Male | 78 | 23 | 55 |
| 28 | 50 |
|
|
Female | 41 | 12 | 29 |
| 16 | 25 |
|
| Tumor location |
|
|
| 0.351 |
|
| 0.906 |
|
Upper | 12 | 3 | 9 |
| 4 | 8 |
|
|
Middle | 77 | 26 | 51 |
| 28 | 49 |
|
|
Lower | 30 | 6 | 24 |
| 12 | 18 |
|
| Tumor size |
|
|
| 0.407 |
|
| 0.113 |
| ≥50
mm | 41 | 10 | 31 |
| 11 | 30 |
|
| <50
mm | 78 | 25 | 53 |
| 33 | 45 |
|
| Differentiation
degree |
|
|
| 0.016 |
|
| 0.001 |
| G1 | 29 | 14 | 15 |
| 19 | 10 |
|
| G2 | 57 | 16 | 41 |
| 17 | 40 |
|
| G3 | 33 | 5 | 28 |
| 8 | 25 |
|
| Local invasion |
|
|
| 0.016 |
|
| 0.004 |
| T1+
T2 | 57 | 23 | 34 |
| 29 | 28 |
|
| T3+
T4 | 62 | 12 | 50 |
| 15 | 47 |
|
| Lymph node
metastasis |
|
|
| 0.002 |
|
| 0.007 |
|
Positive | 71 | 13 | 58 |
| 19 | 52 |
|
|
Negative | 48 | 22 | 26 |
| 25 | 23 |
|
| TNM stage |
|
|
| 0.001 |
|
| 0.002 |
| I +
II | 53 | 24 | 29 |
| 28 | 25 |
|
| III +
IV | 66 | 11 | 55 |
| 16 | 50 |
|
Ku80 expression levels are associated
with prognosis in patients with ESCC
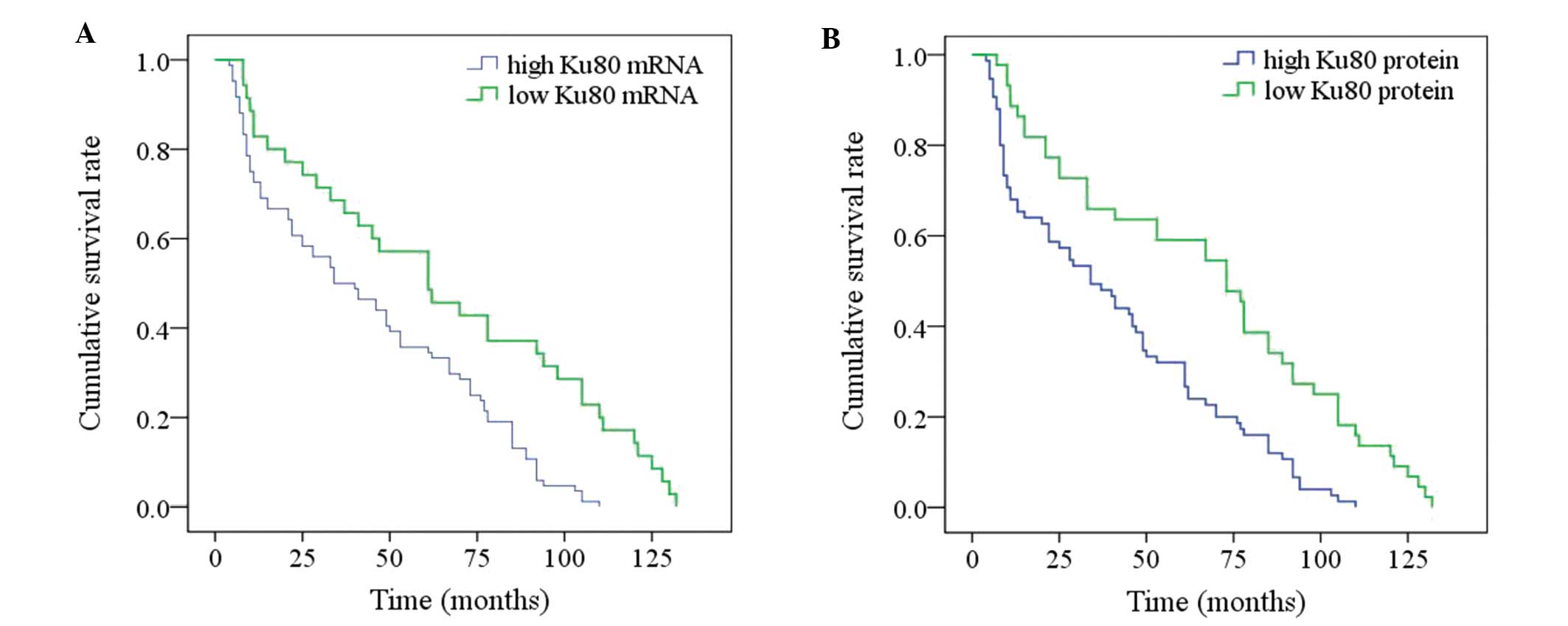
Patients with ESCC were followed during for a median
period of 65.8 months, ranging from 5 to 132 months. The overall
1-, 3-, 5- and 10-year survival rates of the 119 patients were
75.6, 54.6, 42.0 and 5.0%, respectively. The median survival time
was 46.0 months (95% CI, 32.8~59.2 months). As indicated in
Table II and Fig. 3, univariate analysis revealed that
local invasion (P=0.014), lymph node metastasis (P<0.001), TNM
stage (P<0.001) and Ku80 mRNA and protein levels (P<0.001)
were all significant prognostic factors. However, the age of the
patient (P=0.406), gender (P=0.719), tumor location (P=0.248),
tumor size (P=0.339) and differentiation degree (P=0.241) were not
observed to be significant (Table
II). Low aggressive local invasion, negative lymph node
metastasis, early TNM stage and low Ku80 mRNA and protein
expression levels implicated significantly improved prognosis. To
exclude confounding factors, the Cox proportional hazards model was
used to identify factors associated with overall survival of
patients with ESCC. Multivariate analysis revealed that local
invasion (P=0.011), lymph node metastasis (P=0.009), TNM stage
(P<0.001), Ku80 mRNA and protein levels (P=0.024 and 0.007,
respectively) were independent significant prognostic factors.
 | Table II.Univariate and multivariate survival
analyses for patients with ESCC. |
Table II.
Univariate and multivariate survival
analyses for patients with ESCC.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | χ2 | P-value | HR | 95% CI | P-value |
|---|
| Age (years) |
|
|
|
|
|
| ≥60 vs.
<60 | 0.689 | 0.406 | – | – | – |
| Gender |
|
|
|
|
|
| Male
vs. female | 0.129 | 0.719 | – | – | – |
| Tumor location |
|
|
|
|
|
| Upper
vs. middle vs. lower | 2.791 | 0.248 | – | – | – |
| Tumor size
(mm) |
|
|
|
|
|
| ≥50 vs.
<50 | 0.916 | 0.339 | – | – | – |
| Differentiation
degree |
|
|
|
|
|
| G1 vs.
G2 vs. G3 | 2.848 | 0.241 | – | – | – |
| Local invasion |
|
|
|
|
|
| T1+T2
vs. T3+T4 | 6.075 | 0.014 | 1.706 | 1.133–2.568 | 0.011 |
| Lymph node
metastasis |
|
|
|
|
|
|
Positive vs. negative | 29.168 | <0.001 | 1.898 | 1.173–3.073 | 0.009 |
| TNM stage |
|
|
|
|
|
| I+II
vs. III+IV | 26.936 | <0.001 | 2.612 | 1.638–4.163 | <0.001 |
| Ku80 mRNA
levels |
|
|
|
|
|
| Low vs.
high | 13.166 | <0.001 | 1.701 | 1.074–2.694 | 0.024 |
| Ku80 protein
levels |
|
|
|
|
|
| Low vs.
high | 17.169 | <0.001 | 1.765 | 1.164–2.676 | 0.007 |
Discussion
Numerous literature reports have demonstrated
genomic and proteomic changes in ESCC (4–7). However,
the processes underlying the carcinogenesis and progression of ESCC
remain unclear. A previous study indicated that genome instability
through severe DNA damage is associated with the carcinogenesis and
development of ESCC (8). Ku80 is
involved in telomere maintenance, maintenance of chromosomal
integrity, recombination of the V, D and J segments during
immunoglobulin production, regulation of glucose-regulated peptide
78 gene transcription and cell survival (19–22).
Additionally, a number of notable findings regarding the
involvement of the Ku80 gene in cancer have been reported
previously: The overexpression of Ku80 is clearly associated with
the carcinogenesis and the progression of several types of invasive
cancer, including bladder carcinoma (12), gastric carcinoma (13), colorectal carcinoma (23) and breast carcinoma (24). These results suggest that Ku80 may act
as an oncogene in various types of cancer.
In the present study, the role of Ku80 in ESCC was
explored. It was demonstrated that the mRNA expression levels of
Ku80 were increased in ESCC tissues compared with those in CHEM and
NEM (Fig. 1). The RT-qPCR results
were confirmed by IHC, which revealed that the immunohistochemical
scores of Ku80 protein in ESCC were also higher than in CHEM and
NEM. Spearman bivariate correlation analysis indicated that the
expression levels of Ku80 mRNA were positively correlated with Ku80
protein expression levels. This is consistent with the results
reported by Yang et al (11)
and Tonotsuka et al (14), who
demonstrated that Ku80 was overexpressed in ESCC tissues and cell
lines, and suggested that Ku80 may participate in the development
of malignant clinicopathological processes in ESCC.
To further explore the clinicopathological
significance and prognostic value of Ku80, the 119 ESCC patients
were divided into 2 groups of high and low expression, based on the
levels of Ku80 mRNA and protein expression. It is difficult to
determine the extent to which Ku80 expression is pathologically and
clinically relevant, as previous studies had defined the expression
levels of Ku80 using different classification systems (10–12). There
was no universal and commonly accepted classification system, and
the cut-off score was often set arbitrarily. This may lead to
numerous errors in studies aiming to evaluate the association of
Ku80 expression levels with the clinicopathological features of
ESCC. In the present study, an objective, rapid and reproducible
scoring method to set the cut-off score based on a ROC curve
analysis was used. The value closest to the point with maximum
sensitivity and specificity was selected as the cut-off score. By
ROC curve analysis, the largest number of ESCC patients was
correctly classified into different groups using statistical
significance. Thus, there were 84 and 35 patients with ESCC in the
high and low Ku80 mRNA level groups, respectively, and 75 and 44
patients in the high and low Ku80 protein level groups,
respectively. χ2 test indicated that the frequency of
overexpression was significantly higher in ESCC tissues than in
NEM. Additionally, high expression levels of Ku80 were
significantly associated with adverse clinicopathological features,
including low differentiation degree, aggressive local invasion,
lymph node metastasis and advanced TNM stages. Consistent with
these results, the data from the IHC analysis indicated that
increased Ku80 protein expression levels were associated with low
differentiation degree, aggressive local invasion, lymph node
metastasis and advanced TNM stages. However, no correlation was
observed between the Ku80 mRNA or protein expression levels and
several other clinicopathological parameters, including age,
gender, tumor location and tumor size, in the current patient
cohort (Table I). These findings are
consistent with previous reports, which demonstrated that Ku80
expression was associated with important clinicopathological
characteristics in lung adenocarcinoma (25), colorectal cancer (26) and breast cancer (13). These findings suggested that Ku80 was
a bona fide oncogene that may be used as a biomarker and a
therapeutic target in ESCC.
Previous studies evaluating the prognostic
significance of Ku80 expression levels in several types of human
cancer reported different conclusions: In lung adenocarcinoma, Ma
et al (25) demonstrated an
association between Ku80 expression levels and patient outcome in
terms of progression-free survival or overall survival. In
colorectal cancer, Grabsch et al (26) did not observe any association between
high expression levels of Ku80 and long-term survival. In studies
of cases of breast cancer, although Alshareeda et al
(27) have reported an association
with disease-free survival, other authors did not observe such an
association (28). These results may
reflect the complexity of the DNA damage repair mechanisms in
different types of cancer. Therefore, the role of Ku80 in different
pathways and networks and its effect on clinical outcome should be
analyzed in this complex context. In order to investigate whether
Ku80 may be pursued as a novel biomarker for prognostic prediction
in patients with ESCC, the prognosis of 119 patients were analyzed
in the present study using the Kaplan-Meier method. The current
data indicated that the long-term survival of the patients was
associated with low aggressive local invasion, negative lymph node
metastasis, early TNM stage and low Ku80 mRNA and protein
expression levels. Notably, multivariate analysis demonstrated that
local invasion, lymph node metastasis, TNM stage and Ku80 mRNA and
protein expression levels had independent prognostic influence in
patients with ESCC. Therefore, assessment of Ku80 expression in
ESCC may provide valuable information regarding the outcome and
follow-up management.
Nevertheless, there were various limitations to the
present study as follows: Expression of Ku80 in the esophageal
mucosa of the individuals investigated may be differentially
induced by DNA-damage mutagens. Although numerous potential
mutagens, including dietary habits, medicine, alcohol and tobacco
were eliminated, the expression levels of Ku80 are affected by
environmental mutagens to different extents. The disease-associated
alterations in diet and medication were of small concern, since the
patients were diagnosed 6 months prior to the study and did not
receive any therapeutic treatment. Human Ku80 is strictly regulated
by complex transcriptional and translational mechanisms (17,21).
Furthermore, genetic differences among patient cohorts of different
geographical origins may result in differences in Ku80 expression.
One limitation of the present study is that all participants, who
are Han Chinese, come from the same geographical area and have
similar genetic background. Adjuvant chemotherapy or radiotherapy
may also affect the prognosis of patients. Regarding the treatment
modalities, their efficacy was not discussed, since there was no
randomized clinical trial. Another limitation of the present study
is the limited sample size of the controls used to evaluate the
role of Ku80 expression in ESCC. Although, to the best of our
knowledge, this is the first report demonstrating that the
expression of Ku80 is an independent prognostic factor in patients
with ESCC, questions remain unanswered regarding the mechanism of
action of Ku80 during the initiation and progression of ESCC.
Therefore, further studies with larger samples of cases at the
various stages of ESCC are warranted to assess the
clinicopathological significance of Ku80.
In conclusion, the present study provides unique
perspectives regarding the involvement of Ku80 in esophageal
carcinogenesis. The data indicate that Ku80 is overexpressed in a
significant proportion of cases of ESCC, and it is associated with
key clinicopathological features and patient prognosis. In
conclusion, the present study provides evidence about the
unrecognized roles that Ku80 may perform in tumorigenesis, and its
potential use as a novel biomarker and therapeutic target for
ESCC.
References
|
1
|
Trivers KF, Sabatino SA and Stewart SL:
Trends in esophageal cancer incidence by histology, United States,
1998–2003. Int J Cancer. 123:1422–1428. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008. GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shibata-Kobayashi S, Yamashita H, Okuma K,
Shiraishi K, Igaki H, Ohtomo K and Nakagawa K: Correlation among 16
biological factors [p53, p21(waf1), MIB-1 (Ki-67), p16(INK4A),
cyclin D1, E-cadherin, Bcl-2, TNF-α, NF-κB, TGF-β, MMP-7, COX-2,
EGFR, HER2/neu, ER, and HIF-1α] and clinical outcomes following
curative chemoradiation therapy in 10 patients with esophageal
squamous cell carcinoma. Oncol Lett. 5:903–910. 2013.PubMed/NCBI
|
|
5
|
Wang S, Liu H, Wang Z and Chen HX: Effects
of 5-azacytidine on RUNX3 gene expression and the biological
behavior of esophageal carcinoma cells. Mol Med Rep. 9:1259–1265.
2014.PubMed/NCBI
|
|
6
|
Rasool S, Khan T, Qazi F and Ganai BA:
ECRG1 and its relationship with esophageal cancer: A brief review.
Onkologie. 36:213–216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
D'Annoville T, D'Journo XB, Loundou A,
Trousse D, Dahan L, Doddoli C, Seitz JF and Thomas PA: Prognostic
impact of the extracapsular lymph node involvement on disease-free
survival according to the 7th edition of American Joint Committee
on Cancer Staging System. Eur J Cardiothorac Surg. 44:e207–211.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kamangar F, Chow WH, Abnet CC and Dawsey
SM: Environmental causes of esophageal cancer. Gastroenterol Clin
North Am. 38:27–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lieber MR, Gu J, Lu H, Shimazaki N and
Tsai AG: Nonhomologous DNA end joining (NHEJ) and chromosomal
translocations in humans. Subcell Biochem. 50:279–296. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fink LS, Lerner CA, Torres PF and Sell C:
Ku80 facilitates chromatin binding of the telomere binding protein,
TRF2. Cell Cycle. 9:3798–3806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang QS, Gu JL, Du LQ, Jia LL, Qin LL,
Wang Y and Fan FY: ShRNA-mediated Ku80 gene silencing inhibits cell
proliferation and sensitizes to gamma-radiation and mitomycin
C-induced apoptosis in esophageal squamous cell carcinoma lines. J
Radiat Res. 49:399–407. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Groselj B, Kerr M and Kiltie AE:
Radiosensitisation of bladder cancer cells by panobinostat is
modulated by Ku80 expression. Radiother Oncol. 108:429–433. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kruer TL, Cummins TD, Powell DW and
Wittliff JL: Characterization of estrogen response element binding
proteins as biomarkers of breast cancer behavior. Clin Biochem.
46:1739–1746. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tonotsuka N, Hosoi Y, Miyazaki S, Miyata
G, Sugawara K, Mori T, Ouchi N, Satomi S, Matsumoto Y, Nakagawa K,
et al: Heterogeneous expression of DNA-dependent protein kinase in
esophageal cancer and normal epithelium. Int J Mol Med. 18:441–447.
2006.PubMed/NCBI
|
|
15
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumours. 7th. Wiley-Blackwell;
Oxford, UK: 2009
|
|
16
|
Hamilton SR and Aaltonen LA: Pathology and
genetics of tumours of the digestive systemWorld Health
Organization Classification of Tumours. Kleihues P and Sobin LH:
IARC Press; Lyon, France: pp. 237–240. 2000
|
|
17
|
Koike M, Shiomi T and Koike A:
Dimerization and nuclear localization of ku proteins. J Biol Chem.
276:11167–11173. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song Y, Wang Z, Liu X, Jiang W and Shi M:
CCR7 and VEGF-C: Molecular indicator of lymphatic metastatic
recurrence in pN0 esophageal squamous cell carcinoma after
Ivor-Lewis esophagectomy? Ann Surg Oncol. 19:3606–3612. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Finnie NJ, Gottlieb TM, Blunt T, Jeggo PA
and Jackson SP: DNA-dependent protein kinase activity is absent in
xrs-6 cells: Implications for site-specific recombination and DNA
double-strand break repair. Proc Natl Acad Sci USA. 92:320–324.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shrivastav M, Miller CA, De Haro LP,
Durant ST, Chen BP, Chen DJ and Nickoloff JA: DNA-PKcs and ATM
co-regulate DNA double-strand break repair. DNA Repair (Amst).
8:920–929. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gullo C, Au M, Feng G and Teoh G: The
biology of Ku and its potential oncogenic role in cancer. Biochim
Biophys Acta. 1765:223–234. 2006.PubMed/NCBI
|
|
22
|
Liu ES and Lee AS: Common sets of nuclear
factors binding to the conserved promoter sequence motif of two
coordinately regulated ER protein genes, GRP78 and GRP94. Nucleic
Acids Res. 19:5425–5431. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lim JW, Kim H and Kim KH: Expression of
Ku70 and Ku80 mediated by NF-kappa B and cyclooxygenase-2 is
related to proliferation of human gastric cancer cells. J Biol
Chem. 277:46093–46100. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hosoi Y, Watanabe T, Nakagawa K, Matsumoto
Y, Enomoto A, Morita A, Nagawa H and Suzuki N: Up-regulation of
DNA-dependent protein kinase activity and Sp1 in colorectal cancer.
Int J Oncol. 25:461–468. 2004.PubMed/NCBI
|
|
25
|
Ma Q, Li P, Xu M, Yin J, Su Z, Li W and
Zhang J: Ku80 is highly expressed in lung adenocarcinoma and
promotes cisplatin resistance. J Exp Clin Cancer Res. 31:992012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grabsch H, Dattani M, Barker L, Maughan N,
Maude K, Hansen O, Gabbert HE, Quirke P and Mueller W: Expression
of DNA double-strand break repair proteins ATM and BRCA1 predicts
survival in colorectal cancer. Clin Cancer Res. 12:1494–1500. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alshareeda AT, Negm OH, Albarakati N,
Green AR, Nolan C, Sultana R, Madhusudan S, Benhasouna A, Tighe P,
Ellis IO and Rakha EA: Clinicopathological significance of
KU70/KU80, a key DNA damage repair protein in breast cancer. Breast
Cancer Res Treat. 139:301–310. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leifler K Söderlund, Queseth S, Fornander
T and Askmalm MS: Low expression of Ku70/80, but high expression of
DNAPKcs, predict good response to radiotherapy in early breast
cancer. Int J Oncol. 37:1547–1554. 2010.PubMed/NCBI
|