Introduction
Endometrial adenocarcinoma is the most frequently
diagnosed type of gynecological malignancy, with 280,000 cases
reported worldwide in 2008 (1). In
the United States, it is estimated that there were ~40,000 new
cases per year, with 7,500 mortalities reported in 2008 (2). The majority of cases are grade 1
endometrioid-type adenocarcinoma confined to the uterus for which
total hysterectomy and bilateral salpingo-oophorectomy is standard
treatment (1,2). However, a small but significant
percentage of cases recur following total hysterectomy. This
recurrence, or cases that are metastatic upon presentation, exhibit
a limited response to systemic therapy (chemotherapy, hormonal
therapy or radiation). The five-year survival rate is >95% for
women with disease confined to the uterus (stage I) and ~15% for
distant, high-stage disease (3).
Recently, The Cancer Genome Atlas (TCGA) project stratified
endometrioid endometrial adenocarcinoma (EEC) into four different
molecular subgroups, each with distinct survival outcomes: i)
Copy-number low (CNL) alterations; ii) copy-number high alterations
(serous-like); iii) polymerase ε (POLE) ultramutated; and
iv) microsatellite instability hypermutated (4). In this stratification system, the
CNL-EEC molecular subgroup exhibited intermediate progression-free
survival compared with the POLE ultramutated (best
prognosis) and copy-number high, serous-like (worst prognosis)
tumors (4).
Similar to previous studies, which revealed a high
percentage of somatic phosphatase and tensin homolog (PTEN),
phosphoinositide-3-kinase catalytic subunit alpha (PIK3CA)
and PI3K regulatory subunit 1 (PIK3R1) mutations in EECs,
the TCGA study revealed that a significant number of endometrial
adenocarcinomas had genetic alterations in the PI3K/AKT pathway,
suggesting a potential for targeted therapy with specific
inhibitors (4–11). More specifically, TCGA demonstrated
that the PI3K pathway was altered in 92% (83/90) CNL-EEC cases, as
evidenced by somatic mutations in the PTEN, PIK3CA
and PIK3R1 genes (4). The
majority of CNL-EEC tumors (77%; 69/90) exhibited somatic
PTEN mutations, while approximately half (53%; 48/90)
exhibited somatic PIK3CA mutations and one-third (33%;
30/90) contained somatic PIK3R1 mutations. Furthermore,
approximately two-thirds of tumors (68%; 61/90) exhibited somatic
mutations in >1 member of the PI3K pathway (4). These and previous findings (4–11) support
the hypothesis that activation of the PI3K signaling pathway is an
important step in the development of EEC.
The aim of the present study was to investigate
whether mutations of PI3K pathway members are associated with
specific survival outcomes in patients with endometrial
adenocarcinomas and whether these outcomes are specific to a
particular molecular subgroup.
Materials and methods
Survival analysis
The clinical data of 307 newly diagnosed endometrial
carcinoma patients with endometrioid-type histology, including 90
cases of CNL-EEC, was obtained from TCGA Research Network (4) and integrated with PIK3CA mutation
data from TCGA Data Portal Open-Access directory (https://tcga-data.nci.nih.gov/tcga/). Overall and
disease-free survival analysis were then performed using the cBio
Cancer Genomics Portal (http://cbioportal.org). The present study was approved
by the Beth Israel Deaconess Medical Center-HCC Institutional
Review Board.
Review of pathological data
Pathology reports were downloaded from TCGA Data
Portal Open-Access directory or via the cBio Cancer Genomics
Portal. Quality control hematoxylin and eosin (H&E) images from
frozen and permanent sections were re-analyzed by a board-certified
gynecological pathologist (Dr Douglas Lin) at the Department of
Pathology, Beth Israel Deaconess Medical Center (Boston, MA, USA)
to determine that tumors were indeed of the endometrioid
histological subtype. Re-analysis was performed via TCGA BioSig
website (http://tcga.lbl.gov:8080/biosig/tcgadownload.do)
hosted by Lawrence Berkeley National and via the Cancer Digital
Slide Archive (http://cancer.digitalslidearchive.net; Emory
University, Atlanta, GA, USA). Tumor stage (12) and overall International Federation of
Gynecology and Obstetrics (FIGO) grade (13) were also independently re-evaluated
where possible, however, this was deferred to the available
pathology reports, as only limited virtual images from each case
were publicly available for review.
Analysis of PIK3CA mutations
Somatic PIK3CA mutations derived from Next
Generation exome sequencing were retrieved from TCGA Data Portal
Open-Access directory via the cBio Portal (http://www.cbioportal.org/public-portal/). The effect
of specific somatic mutations on PIK3CA function was
determined by cross-referencing them with previously published
studies, wherein the effect of specific PIK3CA mutations on
p110-α activation or gain-of-function was demonstrated in
vitro or in vivo (9,14–16). p110-α is the protein encoded by the
PIK3CA gene and represents the catalytic subunit of PI3K.
The presence of specific PIK3CA somatic mutations in other
cancer types was determined by analyzing the Catalogue of Somatic
Mutations in Cancer database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/).
All websites were accessed between July 1, 2013 and May 1,
2014.
Statistical analysis
Kaplan-Meier overall and disease-free survival
curves and corresponding P-values were calculated using the cBio
Cancer Genomics Portal (http://cbioportal.org; Memorial Sloan Kettering Cancer
Center, New York, NY, USA) with significance estimated by log-rank
test (17,18). Statistical analysis of
clinicopathological characteristics (age and tumor stage and grade)
were assessed using the t-test and paired t-test, respectively.
P<0.05 was considered to indicate a statistically significantly
difference.
Results
Clinical data from 307 newly diagnosed endometrial
carcinoma patients with endometrioid-type histology, including 90
cases of the CNL-EEC subtype, was obtained from TCGA Research
Network (4) and integrated with
PIK3CA mutation data from TCGA Data Portal via the cBio
Cancer Genomics Portal (4,17,18).
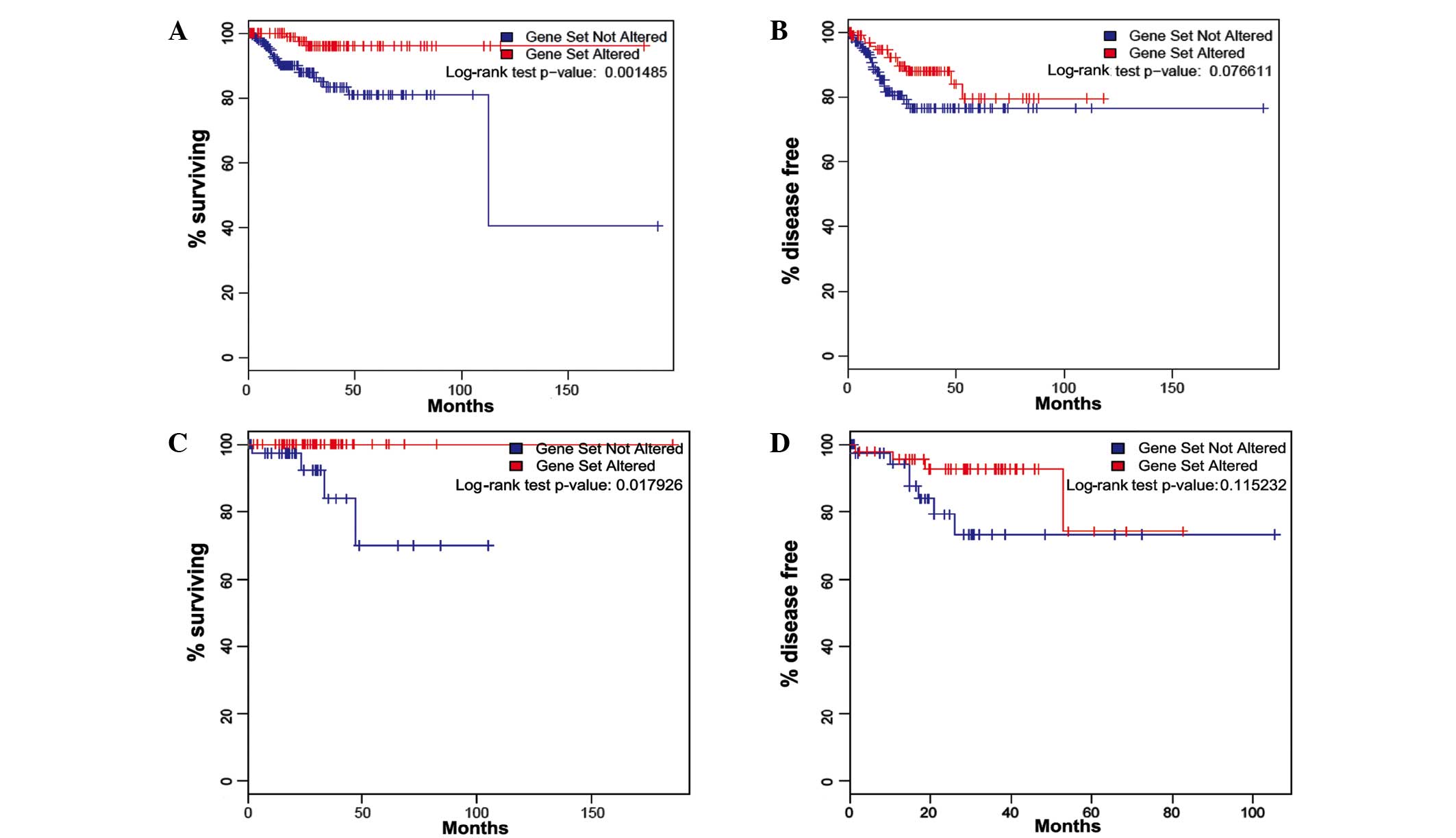
Survival analysis of all TCGA tumors with endometrioid-type
histology (n=307), regardless of molecular subtype, revealed that
patients with somatic PIK3CA mutations exhibited
significantly improved overall survival and marginally prolonged
progression-free survival compared with those possessing wild-type
PIK3CA (log-rank test P=0.001 and P=0.077, respectively;
Fig. 1A and B).
The overall survival outcome difference was specific
to the CNL-EEC patients (n=90; P=0.018; Fig. 1C and D), as no significant association
with overall and disease-free survival was observed in the three
other TCGA molecular subgroups (copy-number high, P=0.442 and
P=0.225; POLE ultramutated, no mortality or recurrence; or
microsatellite instability hypermutated tumors, P=0.108 and
P=0.949) (data not shown). For this reason, the remainder of the
study was focused on the CNL-EEC subgroup. In CNL-EEC, the mean
follow-up times for patients with mutated and wild-type
PIK3CA were 33 and 26 months, respectively. The median
follow-up times for patients with mutated PIK3CA and
wild-type PIK3CA were 29 and 21 months, respectively. There
were no reported mortalities among patients with PIK3CA
mutations (0%; 0/48), while 4 mortalities were reported among the
patients with wild-type PIK3CA (9.5%; 4/42). Of the patients
with somatic PIK3CA mutations, 5 exhibited recurrent disease
(10.4%; 5/48), while 8 patients with wild-type PIK3CA had
recurrent disease (19.0%; 8/42).
Next, the pathological data was re-reviewed in terms
of patient age, tumor type, stage and FIGO grade via the tools
available at TCGA Data Portal Open-Access directory and the cBio
portal. Patient age in PIK3CA-mutated cases ranged from
34–89 years (mean, 60.3 years; median, 60 years), which was similar
to that of the non-mutated cases (range, 37–90 years; mean, 60.6
years; median, 59 years) (Table I).
The majority of PIK3CA-mutated cases were of stage I (83%;
40/48) and FIGO grade 1 or 2 disease (48 and 44%, respectively)
(Table I and Fig. 2). There were no significant
differences in age, tumor stage and FIGO grade between
PIK3CA-mutated and non-mutated cancers (Table I). For all cases in the CNL-EEC
subgroup, the tumors were of endometrioid histological type. This
was confirmed by the presence of back to back atypical endometrial
type glands with complex architecture, no intervening stroma and
varying differentiation upon review of available H&E images
(Fig. 2).
 | Table I.Clinicopathological characteristics of
patients in The Cancer Genome Atlas copy-number low endometrioid
endometrial carcinoma molecular subgroup with and without somatic
PIK3CA mutations. |
Table I.
Clinicopathological characteristics of
patients in The Cancer Genome Atlas copy-number low endometrioid
endometrial carcinoma molecular subgroup with and without somatic
PIK3CA mutations.
|
| PIK3CA |
|
|---|
|
|
|
|
|---|
| Characteristic | Non-mutated | Mutated | P-value |
|---|
| Cases, n | 42 | 48 |
|
| Age, years |
|
| 0.902 |
|
Range | 37–90 | 34–89 |
|
| Mean | 60.6 | 60.3 |
|
|
Median | 59 | 60 |
|
| Stage, n (%) |
|
| 0.406 |
| I | 34 (81.0) | 40 (83.3) |
|
| II | 1 (2.3) | 2 (4.0) |
|
| III | 6
(14.3) | 6
(13.0) |
|
| IV | 1 (2.3) | 0 (0.0) |
|
| Grade, n (%) |
|
| 0.074 |
| 1 | 22 (52.4) | 23 (47.9) |
|
| 2 | 18 (42.9) | 21 (43.8) |
|
| 3 | 2 (4.8) | 4 (8.3) |
|
Pathological analysis of 3 PIK3CA-mutated
cases also demonstrated the presence of a serous carcinoma
component. However, the images that were available for review were
more consistent with endometrioid-type tumors. In addition, these 3
cases exhibited no TP53 mutations or homozygous deletion,
indicating that the histological type was not serous carcinoma.
These findings suggest that survival differences between
PIK3CA mutated and non-mutated cases may not be attributed
to differences in patient age, tumor stage, FIGO grade or
histological subtype.
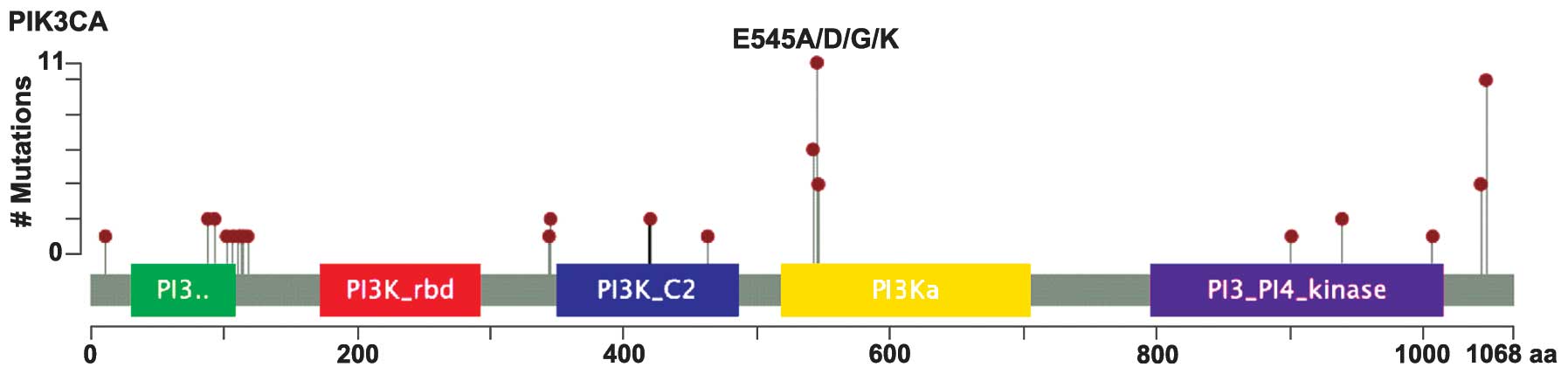
A total of 56 somatic PIK3CA mutations were
identified in 48 patients with CNL-EEC, and a subset of these
mutated tumors (12.5%; 6/48) contained >1 PIK3CA
mutation. PIK3CA mutations occurred predominantly in
hotspots centered on p110-α functional domains (Fig. 3), and around 30% (17/56) occurred
within exons 1–7, which encode the N-terminal domains of p110-α,
including the p85/adaptor-binding domain and the protein kinase-C
homology 2 domain. In addition, around 70% (39/56) of PIK3CA
mutations occurred within exons 9 and 20, which encode the
C-terminal helical and kinase domains (Fig. 3). The majority of cases (79% of
patients and 75% of overall mutations) contained mutations
implicated in PIK3CA activation and gain-of-function
(Table II). The following recurrent
activating mutations were also identified: R88Q, R93Q, N345K,
C420R, E542K, E545K (most common), Q546P, D939G, M1043V, M1043I,
H1047R (second most common) and H1047L (Fig. 3 and Table
II). These findings support the hypothesis that endometrial
adenocarcinomas possess a unique spectrum of somatic PIK3CA
mutations in which a significant number of mutations occur in the
amino terminal domain of p110-α compared with other cancer types,
such as colorectal, breast or bladder carcinomas (4,9).
 | Table II.Summary of somatic PIK3CA
mutations in the copy-number low endometrioid endometrial
adenocarcinoma subgroup. |
Table II.
Summary of somatic PIK3CA
mutations in the copy-number low endometrioid endometrial
adenocarcinoma subgroup.
| Mutation | n | Effect on
function | Type | Domain | Present in other
cancers |
|---|
| WGIHLMPP11del | 1 | Unknown | IF del | ABD | No |
| R88Q | 2 | Activating | Missense | ABD | Yes |
| R93Q | 2 | Activating | Missense | ABD | Yes |
| I102del | 1 | Unknown | IF del | ABD | No |
| N107S | 1 | Unknown | Missense | ABD | No |
| K111E | 1 | Activating | Missense | ABD | Yes |
| L113del | 1 | Unknown | IF del | ABD | Yes |
| R115L | 1 | a | Missense | ABD | Yes |
| G118D | 1 | Activating | Missense | ABD | Yes |
| V344M | 1 | Unknown | Missense | C2 | Yes |
| N345K | 2 | Activating | Missense | C2 | Yes |
| C420R | 2 | Activating | Missense | C2 | Yes |
|
463_465GSN>D | 1 | Unknown | IF del | C2 | No |
| E542K | 5 | Activating | Missense | Helical | Yes |
| E542A | 1 | Activating | Missense | Helical | Yes |
| E545K | 8 | Activating | Missense | Helical | Yes |
| E545A | 1 | Activating | Missense | Helical | Yes |
| E545G | 1 | Activating | Missense | Helical | Yes |
| E545D | 1 | a | Missense | Helical | Yes |
| Q546P | 2 | Activating | Missense | Helical | Yes |
| Q546K | 1 | Activating | Missense | Helical | Yes |
| Q546R | 1 | a | Missense | Helical | Yes |
| C901F | 1 | Unknown | Missense | Kinase | Yes |
| D939G | 2 | Unknown | Missense | Kinase | Yes |
| G1007R | 1 | Unknown | Missense | Kinase | Yes |
| M1043V | 2 | Activating | Missense | Kinase | Yes |
| M1043I | 2 | Activating | Missense | Kinase | Yes |
| H1047R | 7 | Activating | Missense | Kinase | Yes |
| H1047L | 2 | Activating | Missense | Kinase | Yes |
| H1047Q | 1 | a | Missense | Kinase | Yes |
Discussion
Despite its growth-promoting properties, the effect
of PIK3CA mutation on patient prognosis varies according to
the cancer subtype. In patients with colorectal or lung cancer,
mutations of PIK3CA are associated with poor prognosis. By
contrast, PIK3CA mutations have been associated with
favorable prognosis in patients with breast cancer and esophageal
squamous cell carcinomas (19–22). In
endometrial cancer patients, the influence of PIK3CA
mutations on survival and prognosis has been unclear, as there is
data to suggest an association between PIK3CA mutations and
both favorable and unfavorable prognoses (23–25).
The TCGA Data Portal on endometrial adenocarcinoma
offers a host of tools that can be used to interrogate specific
genetic events relative to survival and clinicopathological
characteristics, as well as to generate hypotheses for future
testing. In the present study, we hypothesized that PIK3CA
mutations in EEC may be associated with favorable survival
outcomes, possibly specific to a molecularly sub-defined cohort of
tumors that exhibit low somatic copy-number alterations and
microsatellite stability. The significantly improved overall
survival observed in all patients with endometrioid-type
adenocarcinomas exhibiting PIK3CA mutations, regardless of
the molecular subtype, may reflect the fact that the majority of
TCGA tumors were in the CNL-EEC subgroup.
However, due to the limitations of the TCGA study
(i.e., small sample size of the CNL-EEC subgroup, relatively short
follow-up time, lack of information on co-morbidities), these
findings are exploratory and hypothesis-generating in nature,
rather than definitive. For this reason, future studies with
multivariate analyses and long-term follow-up to determine the
effect of other clinical co-variables (including race, body mass
index, estrogen receptor/progesterone receptor status,
co-morbidities and treatment regimens) on survival outcome are
required. In addition, long-term multivariate analyses with
increased power are essential to determine the effect of other
molecular co-variables. For example, Liang et al (11) previously reported that ARID1A
mutations co-occur with PI3K pathway mutations in endometrial
cancer and lead to activation of the PI3K signaling pathway. In the
TCGA CNL-EEC subgroup, somatic mutations of ARID1A occurred
in 42% of cases (38/90) with a trend towards improved overall
survival (P=0.0517) and no differences in progression-free survival
(P=0.86). By contrast, significantly improved overall survival was
observed in cases with PIK3CA-ARID1A co-occurring
mutations (P=0.00026), with no differences in progression-free
survival (P=0.88; data not shown).
Finally, the mechanism by which PIK3CA
mutations affect patient survival and the mechanism that explains
why there appears to be no association between PIK3R1
mutations and survival outcome are unclear, as PIK3CA and
PIK3R1 are subunits of the same kinase (8,11). One
possible explanation for this is that PIK3CA- and
PIK3R1-mutated tumors may have different driver
mutations.
In conclusion, the results presented in the current
study suggest that PIK3CA mutations may serve as a favorable
prognostic biomarker in EEC, depending on further molecular
sub-stratification of the tumors. Therefore, future studies on
larger independent cohorts with long term follow-up are warranted
to further analyze this association and to validate the findings of
the TCGA project.
Acknowledgements
The present study was performed in accordance with
the The Cancer Genome Atlas (TCGA) publication policy for
endometrial carcinomas, as outlined in TCGA data portal (http://cancergenome.nih.gov). In addition, the current
study has been previously published as an abstract for the 2015
United States and Canadian Academy of Pathology annual meeting
(www.nature.com/modpathol/journal/v27/n2s/pdf/modpathol201415a.pdf).
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RI, De Santis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kandoth C, Schultz N, Cherniack AD, Akbani
R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al:
Cancer Genome Atlas Research Network: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Risinger JI, Hayes AK, Berchuck A and
Barrett JC: PTEN/MMAC1 mutations in endometrial cancers. Cancer
Res. 57:4736–4738. 1997.PubMed/NCBI
|
|
6
|
Oda K, Stokoe D, Taketani Y and McCormick
F: High frequency of coexistent mutations of PIK3CA and PTEN genes
in endometrial carcinoma. Cancer Res. 65:10669–10673. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shoji K, Oda K, Nakagawa S, Hosokawa S,
Nagae G, Uehara Y, Sone K, Miyamoto Y, Hiraike H, Hiraike-Wada O,
et al: The oncogenic mutation in the pleckstrin homology domain of
AKT1 in endometrial carcinomas. Br J Cancer. 101:145–148. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Urick ME, Rudd ML, Godwin AK, Sgroi D,
Merino M and Bell DW: PIK3R1 (p85α) is somatically mutated at high
frequency in primary endometrial cancer. Cancer Res. 71:4061–4067.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rudd ML, Price JC, Fogoros S, Godwin AK,
Sgroi DC, Merino MJ and Bell DW: A unique spectrum of somatic
PIK3CA (p110α) mutations within primary endometrial carcinomas.
Clin Cancer Res. 17:1331–1340. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheung LW, Hennessy BT, Li J, Yu S, Myers
AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, et al:
High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer
elucidates a novel mechanism for regulation of PTEN protein
stability. Cancer Discov. 1:170–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang H, Cheung LW, Li J, Ju Z, Yu S,
Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, et al: Whole-exome
sequencing combined with functional genomics reveals novel
candidate driver cancer genes in endometrial cancer. Genome Res.
22:2120–2129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edge S, Byrd D, Compton C, et al: AJCC
Cancer Staging. Manual (7th). (New York, NY). Springer. 2010.
|
|
13
|
Silverberg SG, Kurman RJ, Nogales F, et
al: Tumors of the uterine corpus: Epithelial tumours and related
conditions. Tumors of the Breast and Gynecologic Tract. Tavassoli
FA and Devilee PL: (Lyon). IARC Press. 2182002.
|
|
14
|
Burke JE, Perisic O, Masson GR, Vadas O
and Williams RL: Oncogenic mutations mimic and enhance dynamic
events in the natural activation of phosphoinositide 3-kinase p110α
(PIK3CA). Proc Natl Acad Sci USA. 109:15259–15264. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gymnopoulos M, Elsliger MA and Vogt PK:
Rare cancer-specific mutations in PIK3CA show gain of function.
Proc Natl Acad Sci USA. 104:5569–5574. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rios JJ, Paria N, Burns DK, Israel BA,
Cornelia R, Wise CA and Ezaki M: Somatic gain-of-function mutations
in PIK3CA in patients with macrodactyly. Hum Mol Genet. 22:444–451.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:112013.
|
|
19
|
Liao X, Morikawa T, Lochhead P, Imamura Y,
Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt
JA, et al: Prognostic role of PIK3CA mutation in colorectal cancer:
Cohort study and literature review. Clin Cancer Res. 18:2257–2268.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Shi L, Zhao X, Wang Y and Yue W:
PIK3CA gene mutation associated with poor prognosis of lung
adenocarcinoma. Onco Targets Ther. 6:497–502. 2013.PubMed/NCBI
|
|
21
|
Kalinsky K, Jacks LM, Heguy A, Patil S,
Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W, et
al: PIK3CA mutation associates with improved outcome in breast
cancer. Clin Cancer Res. 15:5049–5059. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shigaki H, Baba Y, Watanabe M, Murata A,
Ishimoto T, Iwatsuki M, Iwagami S, Nosho K and Baba H: PIK3CA
mutation is associated with a favorable prognosis among patients
with curatively resected esophageal squamous cell carcinoma. Clin
Cancer Res. 19:2451–2459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McIntyre JB, Nelson GS, Ghatage P, Morris
D, Duggan MA, Lee CH, Doll CM and Köbel M: PIK3CA missense mutation
is associated with unfavorable outcome in grade 3 endometrioid
carcinoma but not in serous endometrial carcinoma. Gynecol Oncol.
132:188–193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Catasus L, Gallardo A, Cuatrecasas M and
Prat J: PIK3CA mutations in the kinase domain (exon 20) of uterine
endometrial adenocarcinomas are associated with adverse prognostic
parameters. Mod Pathol. 21:131–139. 2008.PubMed/NCBI
|
|
25
|
Dong Y, Yang X, Wong O, Zhang X, Liang Y,
Zhang Y, Wong W, Nong L, Liao Q and Li T: PIK3CA mutations in
endometrial carcinomas in Chinese women: Phosphatidylinositol
3-kinase pathway alterations might be associated with favorable
prognosis. Hum Pathol. 43:1197–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|