Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer worldwide and develops predominately in individuals
with liver cirrhosis (1). Cirrhosis
is the strongest known risk factor for HCC, particularly cirrhosis
resulting from infection with hepatitis C virus (HCV) or hepatitis
B virus (HBV) (2,3). Additionally, heavy alcohol consumption,
diabetes, obesity and tobacco use have been considered to
contribute to the local burden of HCC (4,5). However,
only a small number of people exposed to these risk factors develop
HCC, suggesting that other environmental and genetic factors may
play a role in HCC development. For this reason, the pathogenesis
of HCC has not been fully elucidated.
Additionally, numerous clinicians rely on
serological α-fetoprotein testing and abdominal ultrasound imaging
for HCC screening (6). However, these
screening tools demonstrate low sensitivity and specificity
(7–9)
and the diagnoses of HCC are made late in the course of the
disease. Therefore, early identification of molecular markers
associated with an increased risk of HCC has been proposed as an
alternative strategy for the diagnosis of HCC.
Epidermal growth factor (EGF) was first isolated in
1962 (10) and plays a critical role
in liver tissue regeneration (11).
In previous years, numerous studies have revealed that the EGF
signaling pathway with the EGF 61A/G polymorphism (rs4444903), a
commonly functional single-nucleotide polymorphism (SNP) in the
5′-untranslated region of the EGF gene, is associated with the risk
of tumorigenesis in multiple human cancers (12–14).
Studies have also reported that the EGF 61A/G polymorphism plays an
important role in the occurrence of liver cancer. At present, there
are three published meta-analyses that have investigated the
association between the EGF 61A/G polymorphism and risk of cancer,
including HCC (15–17). However, none of these studies searched
a sufficient number of published studies and are not limited to
HCC. Therefore, the studies are not conclusive in resolving the
role of the EGF 61A/G polymorphism in HCC. Thus, the present
meta-analysis was performed to address the association between the
frequency of the EGF 61A/G polymorphism and the risk of HCC, and to
complete an in-depth subgroup analysis of the study population
characteristics.
Materials and methods
Inclusion criteria
The present meta-analysis was reported according to
the Preferred Reporting Items for Systematic Reviews and
Meta-Analyses statement (18).
Studies that met all of the following criteria were included: (1)
Use of a cohort or case-control design; (2) sufficient data for
examining an odds ratio (OR), with its 95% confidence interval
(CI); (3) assessment of the EGF 61A/G polymorphism and HCC risk;
and (4) the diagnosis of HCC was confirmed histologically,
pathologically or cytologically. The titles and abstracts of all
relevant studies were evaluated, and case reports, editorials and
reviews were excluded.
Search strategy
All cohort studies and case-control studies of the
EGF 61A/G polymorphism and risk of HCC published prior to May 1,
2014 were identified through systematic searches in the PubMed
(National Institutes of Health, Bethesda, MA, USA) and EMBASE
(Elsevier, Amsterdam, Netherlands) databases, using the following
search strategy: (‘epidermal growth factor’ or ‘EGF’) AND
‘polymorphism’ AND (‘hepatocellular carcinoma’ or ‘liver cancer’ or
‘HCC’). In addition, the reference lists of relevant publications
were manually searched by two independent investigators.
Data extraction
For each study, the first author, year of
publication, ethnicity of the population, type of control, number
of patients and control individuals, genotyping method and Hardy
Weinberg equilibrium (HWE) was extracted for the control group. The
results were compared and discrepancies were resolved by consensus
between two independent investigators.
Statistical analysis
The odds ratios (ORs) and relative 95% confidence
intervals (CIs) were used to assess the strength of associations
between the EGF 61A/G polymorphism and the risk of HCC by comparing
five genetic models, which consisted of the G vs. A, AG vs. AA, GG
vs. AA, GG vs. AG + AA, and AG + GG vs. AA models. Subgroup
analysis was also performed based on the ethnicity and type of
controls. Heterogeneity among the studies used was tested using the
I2 test (19).
I2<40% indicated an acceptable heterogeneity among
the included studies in the present meta-analysis and the
fix-effect model was used, otherwise the random-effect model was
used.
The sensitivity analysis was conducted by omitting
any single included study each turn. Publication bias was assessed
by visual inspection of the funnel plots of the primary outcome and
the Egger's test (20). The funnel
plot was considered to be asymmetrical if the intercept of the
Egger's regression line significantly deviated from zero, with a
P-value of <0.05. HWE in the control group was assessed using
Fisher's exact test, with P<0.05 considered to indicate a
statistically significant difference. All statistical tests for the
present meta-analysis were conducted using Comprehensive
Meta-Analysis software, Version 2.2 (Biostat, Inc., Englewood, NJ,
USA).
Results
Study selection and patient
characteristics
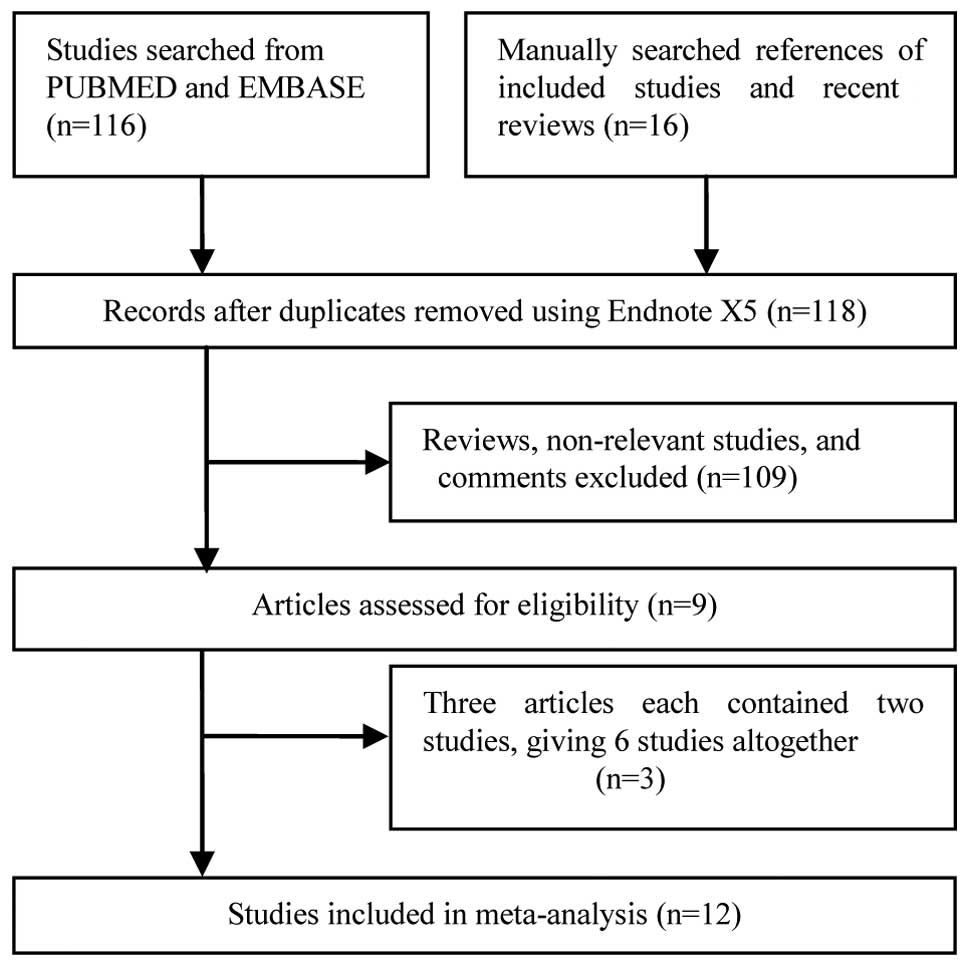
The combined search yielded 132 studies, 123 of
which were excluded as they clearly did not satisfy the inclusion
criteria or were overlapping references (two or more publications
from the same institute or duplicate publication using different
languages). The publications by Zhong et al (15), Tanabe et al (21) and Yuan et al (22) all involved two independent
case-control studies and were overall considered to be six single
studies. Finally, a total of 12 studies (15,21–28) that
examined the association between the EGF 61A/G polymorphism and the
risk of HCC were included in the current meta-analysis (Fig. 1).
A database was created according to the information
extracted from each study. The detailed characteristics of the
included studies are summarized in Table
I. Overall, 2,095 patients with HCC and 3,766 control
individuals were retrieved. Seven of the studies enrolled Chinese
individuals (15,22–26), three
studies involved a mixed population, including Caucasian, Hispanic
and Asian populations and individuals of African descent (21,22,27), one
study enrolled only Caucasians (21)
and one enrolled only Egyptian individuals (28). The genotype distributions in the
controls for all studies were consistent with the HWE
expectations.
 | Table I.Characteristics of included studies in
the meta-analysis. |
Table I.
Characteristics of included studies in
the meta-analysis.
|
|
| Hepatocellular
carcinoma group |
| Control group |
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| First author, year
(ref.) | Ethnicity | Total | GG | AG | AA | Type of control | Total | GG | AG | AA | Genotyping
method | HWE |
|---|
| Tanabe, 2008a
(21) | Mixed | 59 | 23 | 27 | 9 | Cirrhosis | 148 | 32 | 65 | 51 | PCR-RFLP | 0.19 |
| Tanabe, 2008b
(21) | Caucasian | 44 | 15 | 17 | 12 | Cirrhosis | 77 | 12 | 37 | 28 | PCR-RFLP | 0.97 |
| Li, 2009 (25) | Chinese | 186 | 96 | 82 | 8 | Healthy | 186 | 96 | 73 | 17 | PCR-RFLP | 0.56 |
|
|
|
|
|
|
| Cirrhosis | 152 | 65 | 72 | 15 |
| 0.44 |
| Qi, 2009 (24) | Chinese | 215 | 102 | 98 | 15 | Healthy | 208 | 104 | 84 | 20 | PCR-RFLP | 0.64 |
|
|
|
|
|
|
| HBV infection | 172 | 78 | 76 | 18 |
| 0.97 |
| Zhong, 2012a
(15) | Chinese | 397 | 200 | 163 | 34 | Mixed | 480 | 209 | 222 | 49 | PCR-RFLP | 0.37 |
| Zhong, 2012b
(15) | Chinese | 217 | 125 | 76 | 16 | Mixed | 200 | 94 | 89 | 17 | PCR-RFLP | 0.53 |
| Chen, 2011
(26) | Chinese | 120 | 62 | 51 | 7 | Healthy | 120 | 61 | 49 | 10 | PCR-RFLP | 0.97 |
|
|
|
|
|
|
| HBV infection | 120 | 45 | 61 | 14 |
| 0.33 |
| Abu, 2011 (27) | Mixed | 66 | 26 | 25 | 15 | HCV infection | 750 | 180 | 350 | 220 | Real-time PCR | 0.08 |
| Abbas, 2012
(28) | Egyptian | 20 | 7 | 9 | 4 | Healthy | 20 | 2 | 6 | 12 | PCR | 0.37 |
|
|
|
|
|
|
| HCV infection or
cirrhosis | 40 | 7 | 22 | 11 |
| 0.48 |
| Yuan, 2013a
(22) | Mixed | 117 | 28 | 61 | 28 | Healthy | 225 | 63 | 102 | 60 | TaqMan | 0.16 |
| Yuan, 2013b
(22) | Chinese | 250 | 25 | 99 | 126 | Healthy | 245 | 20 | 107 | 118 | TaqMan | 0.53 |
| Wu, 2013 (23) | Chinese | 404 | 206 | 153 | 45 | Healthy | 623 | 291 | 256 | 76 | TaqMan | 0.09 |
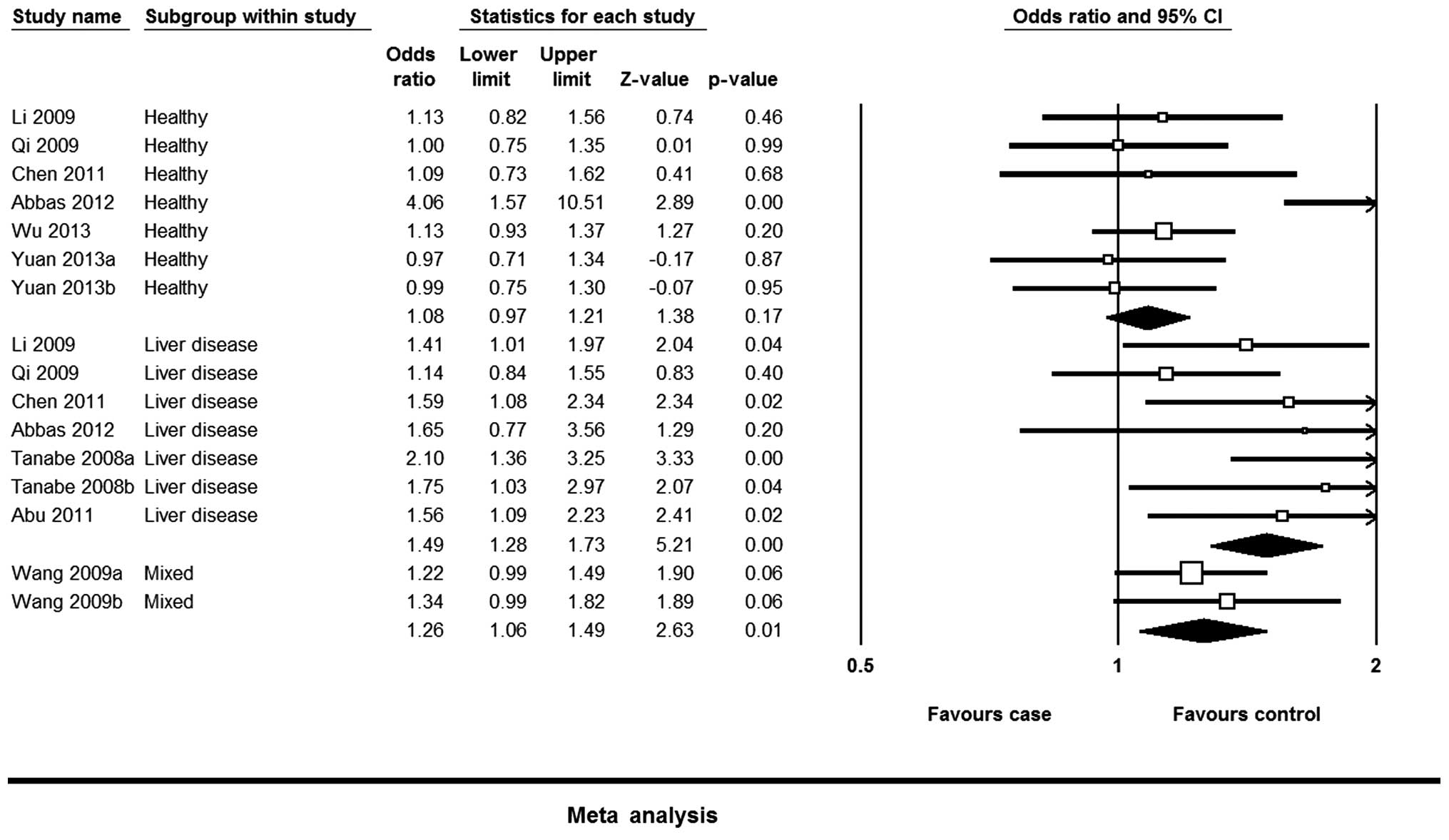
Overall analysis
The evaluation of association between the EGF 61A/G
polymorphism and the risk of HCC is reported in Table II. Calculation of overall ORs in the
total population demonstrated that the EGF 61A/G polymorphism was
associated with increased risk of HCC in the total population in
the G vs. A (OR, 1.25; 95% CI, 1.11–1.40), GG vs. AA (OR, 1.53; 95%
CI, 1.26–1.85), GG vs. AG + AA (OR, 1.34; 95% CI, 1.13–1.58) and GG
+ AG vs. AA (OR, 1.27; 95% CI, 1.08–1.49) models.
 | Table II.Overall and subgroups meta-analysis
of EGF 61A/G polymorphism and HCC risk. |
Table II.
Overall and subgroups meta-analysis
of EGF 61A/G polymorphism and HCC risk.
|
|
| G vs. A | GG vs. AA | AG vs. AA | GG vs. AG+AA | GG+AG vs. AA |
|---|
|
|
|
|
|
|
|
|
|---|
| Characteristic | n | OR (95% CI) | I2,
% | OR (95% CI) | I2,
% | OR (95% CI) | I2,
% | OR (95% CI) | I2,
% | OR (95% CI) | I2,
% |
|---|
| Overall | 12 | 1.25
(1.11–1.40) | 40.29 | 1.53
(1.26–1.85) | 27.77 | 1.15
(0.96–1.36) |
0.00 | 1.34
(1.13-.58) | 40.62 | 1.27
(1.08–1.49) | 14.57 |
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
|
Chinese | 7 | 1.17
(1.06–1.28) |
0.00 | 1.39
(1.11–1.74) |
0.00 | 1.08
(0.88–1.33) | 10.26 | 1.23
(1.08–1.40) |
0.00 | 1.17
(0.97–1.42) |
4.93 |
|
Mixed | 3 | 1.44
(0.93–2.25) | 76.86 | 1.93
(0.87–4.28) | 72.79 | 1.36
(0.93–1.98) | 12.47 | 1.54
(0.79–3.02) | 76.82 | 1.57
(0.96–2.57) | 46.96 |
|
Caucasian | 1 | 1.75
(1.03–2.97) | NA | 2.92
(1.06–8.06) | NA | 1.07
(0.44–2.60) | NA | 2.80
(1.17–6.73) | NA | 1.52
(0.68–3.42) | NA |
|
Egyptian | 1 | 2.18
(1.05–4.50) | NA | 4.47
(1.05–19.07) | NA | 1.85
(0.50–6.79) | NA | 3.05
(0.96–9.74) | NA | 2.49
(0.74–8.36) | NA |
| Source of
controls |
|
|
|
|
|
|
|
|
|
|
|
| Liver
disease | 7 | 1.49
(1.28–1.73) |
0.26 | 3.00
(2.11–4.26) |
0.00 | 1.47
(1.07–2.04) |
0.00 | 1.62
(1.31–1.99) | 26.84 | 1.81
(1.33–2.45) |
0.00 |
|
Healthy | 7 | 1.08
(0.97–1.21) | 31.77 | 1.55
(1.04–2.33) | 46.61 | 1.14
(0.92–1.42) | 35.00 | 1.06
(0.91–1.25) |
0.00 | 1.17
(0.95–1.44) | 38.31 |
|
Mixed | 2 | 1.26
(1.06–1.49) |
0.00 | 1.39
(0.93–2.07) |
0.00 | 1.01
(0.67–1.52) |
0.00 | 1.38
(1.11–1.72) |
0.00 | 1.20
(0.82–1.76) |
0.00 |
Subgroup analysis
The results were similar between the ethnicities,
with the overall results in the Chinese population being similar to
those of the other ethnicities. No significant association was
observed between the EGF 61A/G polymorphism and HCC risk in the
mixed population. When stratifying by source of controls, the EGF
61A/G polymorphism was associated with an increased risk of HCC in
the control individuals with a liver disease. However, the
meta-analysis revealed that there was no association between the
EGF 61A/G polymorphism and the risk of HCC in healthy and mixed
controls (Table II; Fig. 2).
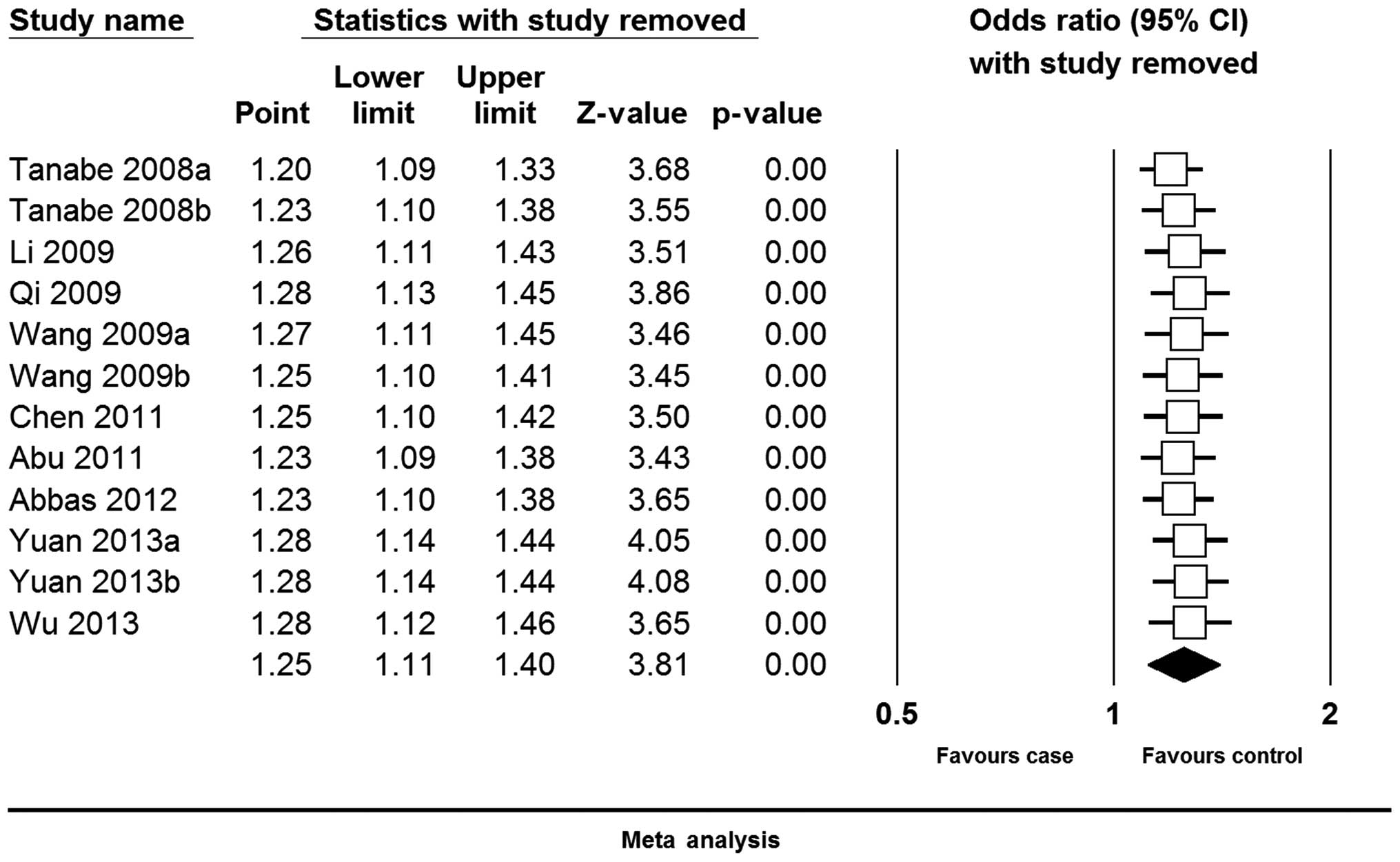
Sensitivity analysis
For the sensitivity analysis, each study involved in
the meta-analysis was omitted each time to reflect the influence of
the individual dataset to the pooled ORs. The corresponding pooled
ORs were not qualitatively altered, indicating that the present
results were statistically robust (Fig.
3).
Publication bias
Funnel plot and Egger's test were performed to
assess the publication bias of literature. The shape of the funnel
plot (Fig. 4) appeared to be
asymmetrical for the EGF 61A/G polymorphism in the genotype
comparison of G vs. A, indicating the presence of publication bias.
Therefore, Egger's test was performed to statistically assess the
symmetry of the funnel plot. The result suggested that publication
bias probably existed in the present study for the G vs. A
(P=0.013), GG vs. AA (P=0.004), AG vs. AA (P=0.011), GG + AG vs. AA
(P<0.001) and GG vs. AG + AA (P=0.051) genotypes.
Discussion
EGF has been hypothesized to promote hepatocyte
transformation, and dysregulation of the EGF signaling pathway has
been speculated to be important in early hepatocarcinogenesis
(29,30). To the best of our knowledge, numerous
previously published genetic studies have demonstrated a positive
association between the EGF 61A/G polymorphism and risk of HCC,
while other studies have found no notable evidence that this
polymorphism increases the susceptibility to HCC. This encouraged
the completion of the present meta-analysis. Meta-analysis is a
method for combining relevant global studies to increase the
statistical power and resolve the discrepancy issue of genetic
association studies (31–34). In the present meta-analysis, a total
of 12 case-control studies involving 2,095 patients and 3,766
control individuals were analyzed to provide a comprehensive
assessment of the association between the EGF 61A/G polymorphism
and HCC risk. The present results for the total population
demonstrated that the EGF 61A/G polymorphism increased the risk of
HCC. In addition, evaluation of heterogeneity was always conducted
in statistical analysis. Thus, the subgroup meta-analyses were
performed according to the ethnicity and source of the control
individuals.
Subsequent to stratification by ethnicity, the
present meta-analysis indicated that the A allele may reduce
susceptibility to HCC in the Chinese population, but not in a mixed
population. This finding in the mixed population is not in
accordance with the results previously published by Zhong et
al (15). In this previous
meta-analysis, a significant association was indicated between the
EGF 61A/G polymorphism and risk of HCC based on eight case-control
studies. The considerably larger sample size of the present study
may account for this difference. The frequency of the AA genotype
varies extensively between different ethnicities, with a prevalence
of 10% in those of Asian descent, ~30% in Caucasians, and 33% in
those of African descent, suggesting a possible ethnicity-based
difference. This may be the reason why no association with the EGF
61A/G polymorphism was detected among the mixed population.
Although environmental factors may be the predominate factors in
the development of HCC, the distribution of EGF genotypes in
various ethnicities may also explain the increased prevalence of
HCC in China (35).
In the stratified analysis by control source, the G
allele was found to be significantly associated with an increased
risk of HCC in the control individuals with liver diseases,
consisting of HBV and HCV infection and cirrhosis. However, there
was no significant association between the EGF 61A/G polymorphism
and the risk of HCC among the healthy control individuals. The
present findings suggest that the EGF 61A/G polymorphism may be a
potential marker in the context of liver disease, consisting of HBV
infection, HCV infection and cirrhosis, rather than a
susceptibility gene polymorphism. The consideration of the history
of relevant diseases was also a strength of the present
meta-analysis compared with the previous meta-analyses performed on
this topic.
There are also limitations to the present study.
First, one of the major concerns is bias, due to selective
publication. Evident publication bias was detected in the G vs. A,
GG vs. AA, AG vs. AA, and GG + AG vs. AA genotype comparisons.
Secondly, the Caucasian and Egyptian populations were assessed in
only one study each, and therefore the results must be interpreted
with caution. Thirdly, the majority of studies were performed using
the Chinese population and additional studies are required using
alternative ethnic groups. Finally, although the heterogeneity in
the present study was not large, it was present in the genetic
models. The subgroup analysis indicated that the heterogeneity may
result from the mixed subgroup. Although heterogeneity is extremely
common in meta-analyses of genetic association, this requires
consideration.
In summary, the present meta-analysis suggests that
the EGF 61A/G polymorphism is associated with an increased risk of
HCC. Based on the evidence obtained in the present meta-analysis,
the EGF 61A/G polymorphism was found to be a potential marker for
HCC in the context of liver disease, such as HBV and HCV infection
and liver cirrhosis. Considering the limited objectives of the
present meta-analysis, additional studies should be conducted with
larger sample sizes and more healthy control designs or prospective
cohort designs.
Acknowledgements
The present study was supported by the Intramural
Evidence-based Medicine Nursery Fund of Taihe Hospital (grant no.,
EBM2013035), funded by the Commonwealth Organization.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Di Costanzo GG: Prospective analysis of
risk factors for hepatocellular carcinoma on patients with
cirrhosis. Hepatology. 38:10612003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thomas MB and Zhu AX: Hepatocellular
carcinoma: the need for progress. J Clin Oncol. 23:2892–2899. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Caldwell SH, Crespo DM, Kang HS and
Al-Osaimi AM: Obesity and hepatocellular carcinoma.
Gastroenterology. 127(Suppl 1): S97–S103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan JM, Govindarajan S, Arakawa K and Yu
MC: Synergism of alcohol, diabetes, and viral hepatitis on the risk
of hepatocellular carcinoma in blacks and whites in the U.S.
Cancer. 101:1009–1017. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy KR: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trevisani F, D'Intino PE, Morselli-Labate
AM, et al: Serum alpha-fetoprotein for diagnosis of hepatocellular
carcinoma in patients with chronic liver disease: Influence of
HBsAg and anti-HCV status. J Hepatol. 34:570–575. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bolondi L, Sofia S, Siringo S, et al:
Surveillance programme of cirrhotic patients for early diagnosis
and treatment of hepatocellular carcinoma: A cost effectiveness
analysis. Gut. 48:251–259. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim CK, Lim JH and Lee WJ: Detection of
hepatocellular carcinomas and dysplastic nodules in cirrhotic
liver: accuracy of ultrasonography in transplant patients. J
Ultrasound Med. 20:99–104. 2001.PubMed/NCBI
|
|
10
|
Cohen S: Isolation of a mouse submaxillary
gland protein accelerating incisor eruption and eyelid opening in
the new-born animal. J Biol Chem. 237:1555–1562. 1962.PubMed/NCBI
|
|
11
|
Michalopoulos GK and DeFrances MC: Liver
regeneration. Science. 276:60–66. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang YM, Cao C and Liang K: Genetic
polymorphism of epidermal growth factor 61A>G and cancer risk: a
meta-analysis. Cancer Epidemiol. 34:150–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lanuti M, Liu G, Goodwin JM, et al: A
functional epidermal growth factor (EGF) polymorphism, EGF serum
levels and esophageal adenocarcinoma risk and outcome. Clin Cancer
Res. 14:3216–3222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Spindler KL, Nielsen JN, Ornskov D,
Brandslund I and Jakobsen A: Epidermal growth factor (EGF) A61G
polymorphism and EGF gene expression in normal colon tissue from
patients with colorectal cancer. Acta Oncol. 46:1113–1117. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhong JH, You XM, Gong WF, et al:
Epidermal growth factor gene polymorphism and risk of
hepatocellular carcinoma: a meta-analysis. PLoS One. 7:e321592012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Z, Wu Q, Shi Y, Nie Y, Wu K and Fan
D: Epidermal growth factor 61A>G polymorphism is associated with
risk of hepatocellular carcinoma: A meta-analysis. Genet Test Mol
Biomarkers. 16:1086–1091. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li TF, Ren KW and Liu PF: Meta-analysis of
epidermal growth factor polymorphisms and cancer risk: Involving
9,779 cases and 15,932 controls. DNA Cell Biol. 31:568–574. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moher D, Liberati A, Tetzlaff J and Altman
DG: PRISMA Group: Preferred reporting items for systematic reviews
and meta-analyses: The PRISMA statement. Ann Intern Med.
151:264–269. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Coory MD: Comment on: Heterogeneity in
meta-analysis should be expected and appropriately quantified. Int
J Epidemiol. 39:932–933. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Egger M, Smith Davey G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tanabe KK, Lemoine A, Finkelstein DM, et
al: Epidermal growth factor gene functional polymorphism and the
risk of hepatocellular carcinoma in patients with cirrhosis. JAMA.
299:53–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yuan JM, Fan Y, Ognjanovic S, et al:
Genetic polymorphisms of epidermal growth factor in relation to
risk of hepatocellular carcinoma: two case-control studies. BMC
Gastroenterol. 13:322013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu J, Zhang W, Xu A, et al: Association of
epidermal growth factor and epidermal growth factor receptor
polymorphisms with the risk of hepatitis B virus-related
hepatocellular carcinoma in the population of North China. Genet
Test Mol Biomarkers. 17:595–600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qi P, Wang H, Chen YM, Sun XJ, Liu Y and
Gao CF: No association of EGF 5′UTR variant A61G and hepatocellular
carcinoma in Chinese patients with chronic hepatitis B virus
infection. Pathology. 41:555–560. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Xie Q, Lu F, et al: Association
between epidermal growth factor 61A/G polymorphism and
hepatocellular carcinoma susceptibility in Chinese patients. Liver
Int. 30:112–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen K, Wei Y, Yang H and Li B: Epidermal
growth factor +61 G/A polymorphism and the risk of hepatocellular
carcinoma in a Chinese population. Genet Test Mol Biomarkers.
15:251–255. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dayyeh Abu BK, Yang M, Fuchs BC, et al: A
functional polymorphism in the epidermal growth factor gene is
associated with risk for hepatocellular carcinoma.
Gastroenterology. 141:141–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abbas E, Shaker O, El Aziz Abd G, Ramadan
H and Esmat G: Epidermal growth factor gene polymorphism 61A/G in
patients with chronic liver disease for early detection of
hepatocellular carcinoma: A pilot study. Eur J Gastroenterol
Hepatol. 24:458–463. 2012.PubMed/NCBI
|
|
29
|
Borlak J, Meier T, Halter R, Spanel R and
Spanel-Borowski K: Epidermal growth factor-induced hepatocellular
carcinoma: Gene expression profiles in precursor lesions, early
stage and solitary tumours. Oncogene. 24:1809–1819. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kömüves LG, Feren A, Jones AL and Fodor E:
Expression of epidermal growth factor and its receptor in cirrhotic
liver disease. J Histochem Cytochem. 48:821–830. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Munafò MR and Flint J: Meta-analysis of
genetic association studies. Trends Genet. 20:439–444. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mao M, Zeng XT, Ma T, He W, Zhang C and
Zhou J: Interleukin-1α-899 (+4845) C→T polymorphism increases the
risk of chronic periodontitis: Evidence from a meta-analysis of 23
case-control studies. Gene. 532:114–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leng WD, He MN, Chen QL, Gong H, Zhang L
and Zeng XT: Vascular endothelial growth factor (VEGF) gene
polymorphisms and risk of head and neck cancer: a meta-analysis
involving 2,444 individuals. Mol Biol Rep. 40:5987–5992. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zeng X, Zhang Y, Kwong JS, et al: The
methodological quality assessment tools for preclinical and
clinical studies, systematic review and meta-analysis, and clinical
practice guideline: A systematic review. J Evid Based Med. 8:2–10.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|