Introduction
Lung cancer is a devastating disease and the leading
cause of cancer-associated mortality worldwide (1). The most frequent type of lung cancer is
non-small-cell lung cancer (NSCLC), which accounts for ~80% of lung
cancer cases (2). The short survival
time of lung cancer patients is mainly attributed to poor outcomes
from conventional chemotherapeutic treatments (3). However, progress in defining the
molecular mechanism of carcinogenesis has led to a notable
improvement in the response to chemotherapy (4). In 2004, it was revealed that epidermal
growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs),
including gefitinib and erlotinib, are only effective in patients
that harbor tumorigenic EGFR mutations that cause aberrant
tyrosine kinase activity (5,6). Thus, identification of oncogenic driver
mutations in cancer patients has become key for the identification
of an effective treatment for NSCLC (7). One of the previously identified
oncogenic driver mutations is the fusion of anaplastic lymphoma
kinase (ALK) with echinoderm microtubule-associated
protein-like 4 (EML4) on chromosome 2p, which was identified
in 2007 in a subpopulation of Japanese patients with NSCLC
(8). In this study, 6.7% (5/75) of
the enrolled Japanese patients with NSCLC possessed EML4-ALK
fusion transcripts, resulting from ALK translocation within
chromosome 2p (8). Other ALK-fusion
genes, including KIF5B-ALK, have also been identified within
chromosome 2p (9–12). Patients possessing ALK fusions
are usually resistant to EGFR-TKIs (13), but respond to the ALK-TKI crizotinib
(14). Therefore, screening for
oncogenic driver mutations, including tumorigenic EGFR
mutations and ALK fusions, has become a crucial step in
disease diagnosis and designing an effective personalized or
tailored therapy plan.
Since the identification of the EML4-ALK
fusion gene, numerous studies have been performed to determine the
frequency of occurrence in patients with NSCLC (8,12,15–24).
However, these numbers varied significantly between studies
(7), ranging between 1.6% in a cohort
of Japanese patients (21) and 11.7%
in a cohort of Chinese patients (22). This is likely to reflect the
differences in detection techniques, sample size and patient
selection criteria. Although the EML4-ALK translocation was
first identified in a NSCLC patient with a history of smoking
(8), subsequent studies have
suggested that the translocation is more frequently detected in
never-smokers (13,16,21,22). A
never-smoker is defined as an individual that has smoked <100
cigarettes per lifetime, according to the US Center for Disease
Control (25). Although inconclusive,
studies have also suggested that the frequency of the incidence is
likely to be increased in female patients compared with male
patients (24). Thus, it is possible
that the frequency of the EML4-ALK translocation may be
markedly higher in female never-smokers. A previous study reported
that the incidence was as high as 15.2% (5/33) in a small cohort of
female patients with adenocarcinoma (24).
To determine the frequency of EML4-ALK fusion
more precisely in female never-smokers, in the present study a
large cohort of patients with NSCLC was assembled. In total, 280
female patients that were never-smokers were enrolled and the
presence of mutations were detected by Multiplex one-step reverse
transcription-polymerase chain reaction (RT-PCR) in the tumor
specimens collected from these patients. The clinical
characteristics that are associated with these mutations were also
analyzed. The present study aimed to increase the understanding of
the EML4-ALK fusion in NSCLC and provide information for
improving the diagnosis procedure and designing personalized
treatment plans.
Materials and methods
Patients and sample collection
The present study was approved by the Institutional
Ethics Committee of Henan Cancer Hospital (Zhengzhou, China). In
total, 280 never-smoking female patients with NSCLC were recruited
(Table I). These patients were
enrolled between 2012 and 2013 at Henan Cancer Hospital. Carcinoma
tissue samples were collected from these patients and preserved as
formalin-fixed paraffin-embedded (FFPE) tissue blocks. The FFPE
tissue blocks were used as the only tissue sources for the
experiments performed in the present study, including the detection
of EML4-ALK fusions and measurement of the expression level
of the ALK tyrosine kinase (ALK TK) mRNA and protein. As a
standardized procedure during diagnosis, tumor subtypes and
pathological characteristics were determined independently by two
pathologists. The opinion of a third pathologist was required when
there was a discrepancy. The tumors were graded based on the
abnormality of the appearance of the tumor cells and tissues
compared to the surrounding normal cells and normal tissues. Tumors
that appeared close to normal were graded as well-differentiated.
Tumors that appeared intermediately and highly abnormal were
classified as moderately- and poorly-differentiated, respectively.
The tumors that appeared to be abnormal were suspected to be
fast-growing and malignant, collectively described as
undifferentiated or less-differentiated tumors in the present
study.
 | Table I.Clinical features of non-small cell
lung cancer tumors in female never-smokers. |
Table I.
Clinical features of non-small cell
lung cancer tumors in female never-smokers.
|
Characteristics | Total, n (%) |
|---|
| Histology |
|
|
Adenocarcinoma | 274 (97.86) |
|
Squamous cell carcinoma | 5
(1.79) |
|
Others | 1
(0.36) |
| Age |
|
| <40
years | 4
(1.43) |
| 40–49
years | 57
(20.36) |
| 50–59
years | 105 (37.50) |
| ≥60
years | 114 (40.71) |
|
Differentiation |
|
|
Poorly-differentiated | 91
(32.50) |
|
Moderately-differentiated | 95
(33.93) |
|
Well-differentiated | 94
(33.57) |
Detection of EML4-ALK fusion
genes
The FFPE tissue blocks were sliced to a width of 3
µm, and the tumor regions were identified and collected for RNA
extraction. Total RNA was extracted using RNeasy FFPE kit (Qiagen,
Valencia, CA, USA), according to the manufacturer's instructions,
and treated with DNase I (DNA-free; Ambion Life Technologies,
Carlsbad, CA, USA) to remove any DNA contamination. The RNA samples
were then subjected to Multiplex One-step RT-PCR with fluorescent
RT-PCR to detect EML4-ALK fusion transcripts using the human Lung
Cancer Related Fusion Gene Detection kit (Yuanqi Bio-Pharmaceutical
Co., Ltd., Shanghai, China), according to the manufacturer's
instructions. Briefly, the mixture of each reaction contained 3 µl
total RNA, 20 µl Multiplex RT-PCR buffer and 2 µl Multiplex Enzyme
Mix in a total volume of 25 µl. The primers included in the
reaction for the detection of EML4-ALK fusion subtype variants were
as follows: V1 and V6 forward, 5′-ATT TGT GCA GTG TTT AGC ATT C-3′;
V2 forward, 5′-CGG GAG ACT ATG AAA TAT TGT ACT-3′; V3a and V3b
forward, 5′-AGT CAC ATA ATT CTT GGG AA-3′; V4b and V7 forward,
5′-GGG AAA GGA CCT AAA GGT G-3′; V4a forward, 5′-GTA GCA GAA GGA
AAG GCA GAT C-3′; V5a and V5b forward, 5′-GCT AAA GGC GGC TTT GGC
TG-3′; and E17-A20 and V9 forward, 5′-CGC TAC TCA ATA GAT GGT ACC
T-3′; and the common reverse primer ALK-E20, 5′-CAT GAT GGT CGA GGT
GCG C-3′ (Sangon Biotech Co., Ltd., Shanghai, China). RT-PCR was
performed using the 7300 Real Time PCR System (Applied Biosystems
Life Technologies, Foster City, CA, USA) under the following
conditions: 42°C for 30 min; 94°C for 5 min; 40 cycles at 94°C for
15 sec; and 60°C for 1 min. The PCR products were subjected to DNA
sequencing (sequencing primer, 5′-TTG CTC AGC TTG TAC TCA GGG CTC
TG-3′; Sangon Biotech Co., Ltd.) to determine the variant types of
the EML4-ALK fusion transcripts. All patients expressing the
EML4-ALK fusion, as determined by Multiplex RT-PCR and
direct DNA sequencing, were further confirmed by Vysis ALK Break
Apart fluorescence in situ hybridization (FISH)
analysis.
ALK break apart FISH analysis
FISH experiments were performed using FFPE tissue
sections in order to identify ALK rearrangements. The Vysis ALK
Break Apart FISH Probe kit (Abbott Molecular Inc., Des Plaines, IL,
USA) was used according to the manufacturer's instructions.
Briefly, two DNA probes that targeted sequences prior to and
following the ALK breaking point were labeled with green and red
florescent dye, respectively, and used for FISH. In normal nuclei
without ALK rearrangement, the red and green fluorescent signals
from the two probes colocalize to form a yellowish signal, or the
two signals are less than two signal diameters apart. However, when
ALK rearrangement occurs, the signals from the two probes are
located more than two signal diameters apart in a single nucleus,
which indicates a cell with ALK rearrangement. A tumor sample was
considered to lack ALK rearrangement if <5 cells out of 50
(<5/50 or <10%) appeared positive for ALK rearrangement. If
>25 cells out of 50 (>25/50 or >50%) were positive for ALK
rearrangement, the tumor was considered positive, but the tumor is
equivocal if 5–25 cells (10 to 50%) are positive for ALK
rearrangement. When the tumor was equivocal, additional evaluation
was required, which was performed according to the manufacturer's
instructions.
Determining the level of ALK mRNA
One-step RT-PCR was performed on the total RNA
extracted from FFPE tissue blocks to determine the level of ALK RNA
in the tissues. Primers specific to the ALK TK domain were designed
for the ALK mRNA expression analysis. The expression level of the
reference gene Abelson murine leukemia viral oncogene homolog (ABL)
was also determined as a control. The primers used were as follows:
ALK forward, 5′-AGA AAC TGC CTC TT GAC CTG-3′ and reverse, 5′-GGG
CAT CCA CTT AAC TGG C-3′; and ABL forward, 5′-TAC CTG AGG GAG TGC
AAC C-3′ and reverse, 5′-TTT TCT TCT CCA GGT ACT CCA-3′. In
addition, the DNA sequencing primers were as follows: ALK
sequencing primer, 5′-CCC TTT CTA TAG TAG CTC GCC CTG TAG AT-3′;
and ABL sequencing primer, 5′-CCA TGT ACA GCA GCA CCA CGG CGT-3′.
RT-PCR was performed as aforementioned.
Immunohistochemistry (IHC)
IHC was used to evaluate the expression of ALK TK
protein, as previously described (26). Briefly, the experiments were performed
on FFPE tissue sections using a Ventana ALK (D5F3) Cdx Assay
(rabbit monoclonal antibody against ALK; cat no. 790–4796; Ventana
Medical Systems, Inc., Tucson, AZ, USA), which detects the
endogenous levels of total ALK protein and ALK fusion proteins,
according to the manufacturer's instructions. The tissue sections
were deparaffinized and incubated with 3%
H2O2 (Ventana Medical Systems, Inc.) to
quench endogenous peroxidase activity, which was followed by
heat-induced antigen retrieval for 30–60 min (Ventana Medical
Systems, Inc.). Subsequent to blocking with 10% normal goat serum
(Ventana Medical Systems, Inc.), the aforementioned primary
antibody were applied, followed by goat anti-rabbit secondary
antibody conjugated with horseradish peroxidase (ultraView
Universal DAB Detection kit; cat no. 760–500; Ventana Medical
Systems, Inc.), according to the manufacturer's instructions. The
immunosignals were visualized using diaminobenzidine (UltraView;
Ventana Medical Systems, Inc.) and counterstained with hematoxylin
(Ventana Medical Systems, Inc.). Images were captured to assess the
intensity of staining, as follows: 0, no staining; 1+, light
staining; 2+, moderate staining; and 3+, strong staining. The
distribution of ALK immunostaining was also determined. Any tissue
specimens exhibiting IHC intensity >0 were defined as IHC
positive.
Statistical analysis
The differences between data were analyzed using
χ2 test, Mann-Whitney U test or Student's
t-test, as appropriate. P<0.05 was considered to indicate
a statistically significant difference.
Results
Clinical features of participating
patients
To avoid the variation caused by a small sample size
and to more precisely determine the frequency of EML4-ALK
translocation, a total of 280 patients with NSCLC were recruited
for the present study. All patients were female and were defined as
never-smokers (25). Based on the
information collected from these patients, the majority of the
patients were diagnosed with adenocarcinoma (97.86%; Table I). The median age of this cohort of
patients was 56.69 years, ranging between 23 and 76 years. Out of
these patients, 40.71% of patients were aged ≥60 years, while
37.50, 20.36 and 1.43% of patients were aged 50–59, 40–49 and
<40 years, respectively (Table I).
The number of patients with poorly, moderately or
well-differentiated tumor cells was similar between the groups,
with 91 (32.50%), 95 (33.93%) and 94 (33.57%) patients in each
group, respectively (Table I).
Detection and characterization of
EML4-ALK translocation
In the present study, the frequency of EML4-ALK
translocation in female never-smokers with NSCLC was determined.
Therefore, Multiplex one-step RT-PCR was performed on the total RNA
extracted from the FFPE tissue samples prepared from the resected
tumors of the enrolled patients. Out of the 280 carcinoma
specimens, EML4-ALK fusion gene transcripts were detected in 21
tissue specimens, accounting for 7.5% of the total patients. The
representative PCR products for each detected EML4-ALK fusion
variant are shown in Fig. 1A. The
presence of ALK rearrangements were confirmed using the Vysis ALK
Break Apart FISH Probe kit, which detects ALK rearrangements using
two fluorescence-labeled probes that target sequences flanking the
ALK breaking point. Representative images of FISH are shown in
Fig. 1B. ALK rearrangements,
indicated by red and green fluorescent signals located at least two
signal diameters apart in a single nucleus, were detected in
carcinoma samples harboring EML4-ALK translocation, but not in
carcinoma samples without EML4-ALK fusion. The identity of these
RT-PCR products was confirmed by DNA sequencing. All RT-PCR
products from 21 tissue samples harboring EML4-ALK translocation
were subsequently sequenced. The DNA sequencing traces and exon
organization for each representative variant are presented in
Fig. 2A. In total, 5 different
variants were detected, including variant 1 in 12 samples and
variant 3a/3b in 6 samples. In addition, variants 4, 5a and E17-A20
were detected in one sample each (Fig.
2B). Variant 2 was not detected in any of the present tissue
samples. Therefore, it is possible that EML4-ALK variant 1,
which accounted for 57.1% of all EML4-ALK translocations, is the
most common form of the fusion products in female never-smokers
with NSCLC. By contrast, the variant 2 is less likely to be
detected in the same group of patients.
 | Figure 1.Screening for EML4-ALK fusion
transcripts in the tissues obtained from female never-smokers with
non-small cell lung cancer. (A) Representative DNA gel images
revealing the expression of the EML4-ALK fusion transcript
variants V1, V3a, V3b, V4a, V5a and E17-20, and ABL controls, as
determined by Multiplex one-step reverse transcription-polymerase
chain reaction. (B) ALK Break Apart fluorescence in situ
hybridization analysis revealed ALK inversion in carcinoma
tissues. Two probes targeting the sequences prior to and following
the breaking point were labeled by red and green fluorescent dye,
respectively. Representative images of EML4-ALK-negative
(left) and -positive (right) carcinomas are shown. The ALK
inversion in EML4-ALK-positive carcinomas is indicated by
two separated red and green dots (right) located at least two
signal diameters apart. NC, negative case; M, DNA ladder;
EML4-ALK, echinoderm microtubule-associated protein-like
4-anaplastic lymphoma kinase; ABL, Abelson murine leukemia viral
oncogene homolog. |
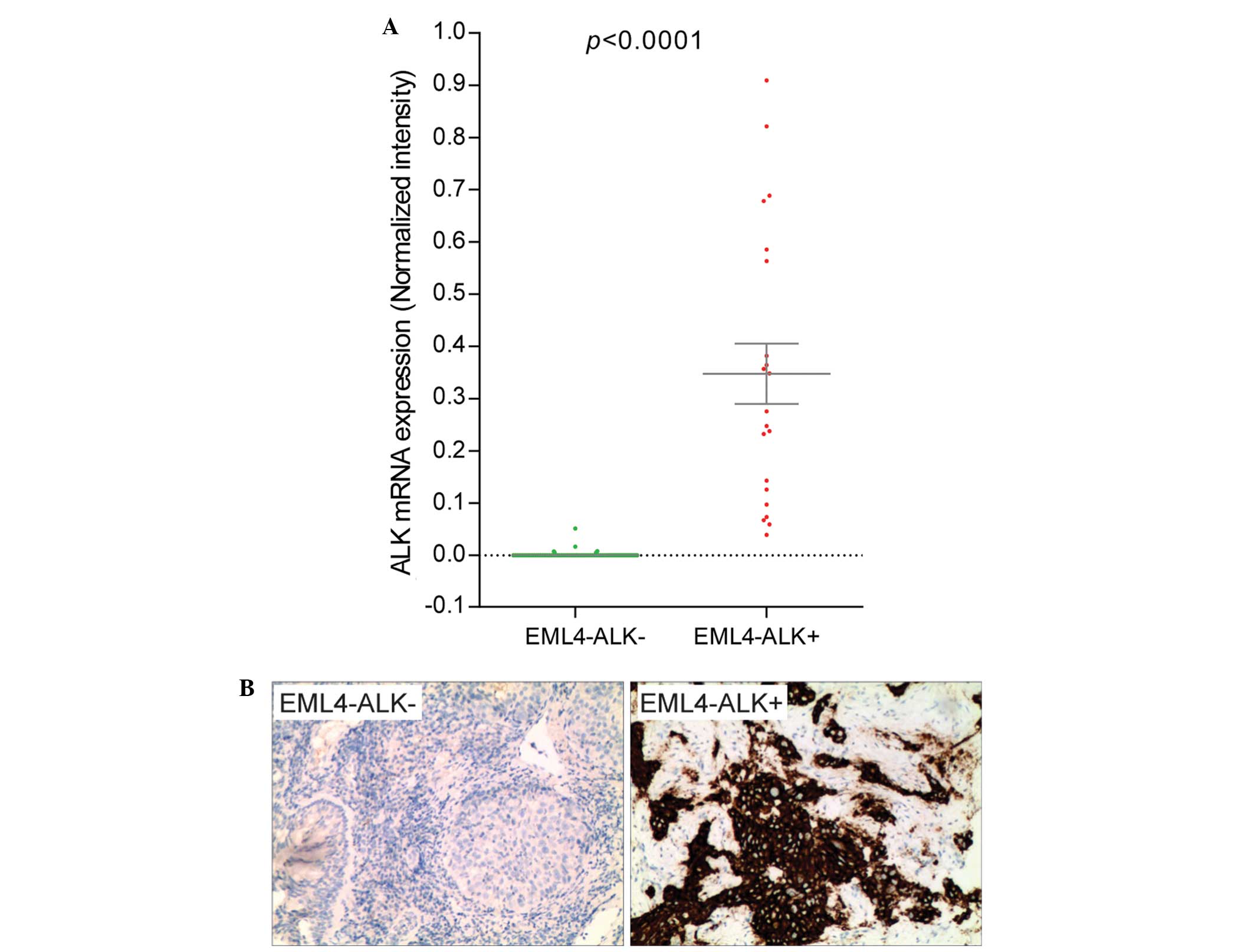
Aberrant expression of ALK mRNA is
associated with EML4-ALK translocation
The expression level of ALK mRNA was measured by
one-step RT-PCR. The primers were specific to the ALK TK
domain that was translocated and fused with EML4 in EML4-ALK fusion
genes, so the mRNAs transcribed from the intact and translocated
ALK genes would be detected. This method revealed that ALK TK mRNA
was highly expressed in tissues harboring EML4-ALK fusion, but not
in tissues without the EML4-ALK fusion (Fig. 3A). Despite expressing different
variants of the EML4-ALK fusion gene, all carcinoma specimens
harboring the fusion expressed ALK TK RNA aberrantly. By contrast,
ALK TK mRNA was not detectable in the majority of the samples not
harboring EML4-ALK translocation. IHC also revealed that the ALK
protein is highly expressed in specimens harboring EML4-ALK fusion,
but the protein was not expressed or was expressed at an extremely
low level in carcinomas without EML4-ALK fusion (Fig. 3B).
EML4-ALK translocation is more
frequently detected in younger patients and undifferentiated
carcinomas
All patients harboring the EML4-ALK translocation in
the present cohort of patients were diagnosed with adenocarcinoma
(Table II). However, it is possible
that the number of other types of carcinoma was too low for
EML4-ALK fusion genes to be detected (6 out of 280 patients;
2.14%). Therefore, the frequency of EML4-ALK translocation in other
types of carcinomas was unclear. In total, 50, 12.28, 7.62 and
3.51% of the patients aged <40, 40–49, 50–59 and ≥60 years,
respectively, harbored EML4-ALK fusion genes (Table II). The distribution of the patients
with EML4-ALK fusion in the four different age groups was
significantly different from the distribution of patients without
EML4-ALK fusion (P<0.0019; Table
II). Furthermore, patients harboring EML4-ALK translocation
(median age, 50.95±2.29 years) were significantly younger than
patients without EML4-ALK translocation (median age, 57.15±0.56;
P<0.01) and all patients combined (median age, 56.69±0.54;
P<0.05) (Fig. 4). Histological
examination revealed that all carcinomas harboring EML4-ALK fusion
were undifferentiated, either poorly- or moderately-differentiated,
but never well-differentiated (Table
II). The difference in the differentiation level between
carcinomas harboring EML4-ALK fusion and those without the fusion
gene is significant (P<0.0014). Overall, these results indicate
that the EML4-ALK fusion is more frequent in younger patients and
in undifferentiated or less-differentiated carcinomas.
 | Table II.Clinical features of tissues
harboring the EML4-ALK fusion gene in female never-smokers
with non-small cell lung cancer. |
Table II.
Clinical features of tissues
harboring the EML4-ALK fusion gene in female never-smokers
with non-small cell lung cancer.
|
| EML4-ALK
fusion gene |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Present, n (%) | Absent, n (%) | Total, n (%) | P-value |
|---|
| Histology |
|
|
|
|
|
Adenocarcinoma | 21 (7.66) | 253 (92.34) | 274 (100) |
|
|
Squamous cell carcinoma | 0
(0.00) |
5
(100.00) |
5 (100) |
|
|
Other | 0
(0.00) |
1
(100.00) |
1 (100) |
|
| Patient age |
|
|
|
|
| <40
years |
2 (50.00) |
2 (50.00) |
4 (100) | 0.0019 |
| 40–49
years |
7 (12.28) | 50
(87.72) | 57
(100) |
|
| 50–59
years | 8
(7.62) | 97
(92.38) | 105 (100) |
|
| ≥60
years | 4
(3.51) | 110 (96.49) | 114 (100) |
|
|
Differentiation |
|
|
|
|
|
Poorly-differentiated | 8
(8.79) | 83
(91.21) | 91
(100) | 0.0014 |
|
Moderately-differentiated | 13
(13.68) | 82
(86.32) | 95
(100) |
|
|
Well-differentiated | 0
(0.00) | 94
(100.00) | 94
(100) |
|
Discussion
By combining multiple published studies, the
frequency of the EML4-ALK translocation in NSCLC worldwide
was determined to be ~5% (7). In the
present study, the frequency of EML4-ALK fusion determined
using Multiplex one-step RT-PCR was 7.5% (21 out of 280 patients)
in a large cohort of never-smoking female patients with NSCLC. This
result was verified by DNA sequencing, ALK Break Apart FISH
analysis, and immunohistochemistry. This frequency is slightly
increased compared with all NSCLC patients worldwide, regardless of
gender and smoking history, indicating that the combined effect of
gender (female) and smoking habit (never-smokers) is minor.
Notably, the present results vary from the findings of a previous
study that assessed 33 female never-smoker patients with NSCLC and
reported that the incidence of EML4-ALK fusion was 15.2% (5
patients) (24). The possible
discrepancy may be due to the variance caused by the difference in
sample sizes between the two studies, with 33 female non-smoking
patients in the previous study and 280 in the present study. In
addition, environmental factors, such as local air pollution levels
and food choices may vary between the different regions of China
and may also be involved in the discrepancy.
Out of the 21 patients with EML4-ALK fusion
genes, more than one-half were detected as fusion gene variant 1.
Variant 2 was not detected in the present tissue samples. It is
therefore possible that variant 1 is the predominant type, while
variant 2 is the least likely to be detected in female
never-smokers with NSCLC. Regardless of the variant present, the
ALK mRNA and protein were aberrantly expressed in all
EML4-ALK-positive carcinomas (Fig.
3). This result is consistent with the findings that
EML4-ALK fusion genes are carcinoma driver mutations
(8) and that overactivation of ALK TK
plays a pivotal role in tumor cell proliferation (26–28).
Compared with conventional chemotherapy, crizotinib, a selective
inhibitor of ALK, has demonstrated a superior ability to improve
the treatment outcome and survival rate in patients harboring
EML4-ALK fusion mutations in clinical trials (26,29–32). By
contrast, these patients demonstrated resistance to EGFR TKIs
(13). Therefore, identification of
EML4-ALK fusion has a direct impact on disease treatment in
NSCLC, particularly with female never-smokers that demonstrate a
high frequency of the EML4-ALK fusion.
In the present cohort of patients, EML4-ALK
fusions were only detected in undifferentiated or
less-differentiated carcinomas, including those graded as poorly-
and moderately-differentiated tumors, but not in tumors graded as
well-differentiated (Table II). This
association between EML4-ALK fusion genes and
undifferentiated carcinomas was also reported in a previous study,
which was conducted using a cohort consisting of male and female
patients with NSCLC (33). Since ALK
is aberrantly expressed only in the carcinoma lesions harboring
EML4-ALK gene fusion, but not in carcinomas without the
fusion gene, it is possible that ALK expression plays an important
role in determining the fate of cells in NSCLC and is accountable
for decreased differentiation, a feature that is often associated
with more rapid-growing and malignant types of carcinomas.
In the present study, the median age of patients
with EML4-ALK gene fusion (50.95 years) was >6 years
younger than the median age of patients without EML4-ALK
gene fusion (57.15 years) in the present cohort of female
never-smokers with NSCLC. The difference in age distribution
between the patients harboring EML4-ALK gene fusion and
those without gene fusion was statistically significant, as
determined by χ2 (P<0.05; Table II). In addition, the frequency of
EML4-ALK gene fusion in the patients aged <40 years is
50%, while the frequency gradually decreases in the older age
groups (Table II). In particular,
only 3.5% of patients aged ≥60 years harbored the EML4-ALK
fusion gene. These results suggest that the age of disease onset is
significantly younger in patients harboring EML4-ALK gene
fusion compared with patients that did not harbor gene fusion.
In conclusion, the frequency of the EML4-ALK
fusion is 7.5% in a cohort of 280 NSCLC patients that consisted of
female never-smokers. Among the identified fusion variants, variant
1 was the most common type, accounting for 57.1% of all
EML4-ALK-positive cases. The mRNA and protein of ALK were
aberrantly expressed in all EML4-ALK-positive carcinoma
lesions, but were absent or expressed at a low level in patients
without the fusion gene, suggesting an important role of
EML4-ALK translocation in tumorigenesis. The EML4-ALK
translocation is detected more frequently in younger patients and
in undifferentiated carcinomas. These results may provide useful
insights to the knowledge of NSCLC and facilitate the diagnosis and
targeted treatment of the disease, leading to an improved treatment
outcome and patient life quality.
Acknowledgements
This study was supported by the Henan Province
Science and Technology Department (grant no., 132300410448).
Abbreviations:
|
ALK
|
anaplastic lymphoma kinase
|
|
EML4
|
echinoderm microtubule-associated
protein-like 4
|
|
NSCLC
|
non-small-cell lung cancer
|
|
EGFR
|
epidermal growth factor receptor
|
|
TKI
|
tyrosine kinase inhibitor
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith W and Khuri FR: The care of the lung
cancer patient in the 21st century: A new age. Semin Oncol. 31(2
Suppl 4): 11–15. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schiller JH, Harrington D, Belani CP,
Langer C, Sandler A, Krook J, Zhu J and Johnson DH: Eastern
Cooperative Oncology Group: Comparison of four chemotherapy
regimens for advanced non-small-cell lung cancer. N Engl J Med.
346:92–98. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Petty RD, Nicolson MC, Kerr KM,
Collie-Duguid E and Murray GI: Gene expression profiling in
non-small cell lung cancer: From molecular mechanisms to clinical
application. Clin Cancer Res. 10:3237–3248. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatulas, Okimoto RA, Brannigan BW, Harris PL, Haserlats M,
Supko JG, Haluska FG, et al: Activating mutations in the epidermal
growth factor receptor underlying responsiveness of non-small-cell
lung cancer to gefitinib. N Engl J Med. 350:2129–2139. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sasaki T, Rodig SJ, Chirieac LR and Jänne
PA: The biology and treatment of EML4-ALK non-small cell lung
cancer. Eur J Cancer. 46:1773–1780. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, et al: Identification of the transforming EML4-ALK
fusion gene in non-small-cell lung cancer. Nature. 448:561–566.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kohno T, Ichikawa H, Totoki Y, Yasuda K,
Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, et
al: KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 18:375–377.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wong DW, Leung EL, Wongs K, Tin VP, Sihoe
AD, Cheng LC, Au JS, Chung LP and Wong MP: A novel KIF5B-ALK
variant in nonsmall cell lung cancer. Cancer. 117:2709–2718. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takeuchi K, Choi YL, Togashi Y, Soda M,
Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y, et al:
KIF5B-ALK, a novel fusion oncokinase identified by an
immunohistochemistry-based diagnostic system for ALK-positive lung
cancer. Clin Cancer Res. 15:3143–3149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rikova K, Guo A, Zeng Q, Possemato A, Yu
J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al: Global survey
of phosphotyrosine signaling identifies oncogenic kinases in lung
cancer. Cell. 131:1190–1203. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shaw AT, Yeap BY, Mino-Kenudson M,
Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S,
McDermott U, et al: Clinical features and outcome of patients with
non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol.
27:4247–4253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thunnissen E, Bubendorf L, Dietel M,
Elmberger G, Kerr K, Lopez-Rios F, Moch H, Olszewski W, Pauwels P,
Penault-Llorca F, et al: EML4-ALK testing in non-small cell
carcinomas of the lung: A review with recommendations. Virchows
Arch. 461:245–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Inamura K, Takeuchi K, Togashi Y, Nomura
K, Ninomiya H, Okui M, Satoh Y, Okumura S, Nakagawa K, Soda M, et
al: EML4-ALK fusion is linked to histological characteristics in a
subset of lung cancers. J Thorac Oncol. 3:13–17. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koivunen JP, Mermel C, Zejnullahu K,
Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas
R, et al: EML4-ALK fusion gene and efficacy of an ALK kinase
inhibitor in lung cancer. Clin Cancer Res. 14:4275–4283. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shinmura K, Kageyama S, Tao H, Bunai T,
Suzuki M, Kamo T, Takamochi K, Suzuki K, Tanahashi M, Niwa H, et
al: EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-,
or TFG-ALK fusion transcripts, in non-small cell lung carcinomas.
Lung Cancer. 61:163–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin E, Li L, Guan Y, Soriano R, Rivers CS,
Mohan S, Pandita A, Tang J and Modrusan Z: Exon array profiling
detects EML4-ALK fusion in breast, colorectal and non-small cell
lung cancers. Mol Cancer Res. 7:1466–1476. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martelli MP, Sozzi G, Hernandez L,
Pettirossi V, Navarro A, Conte D, Gasparini P, Perrone F, Modena P,
Pastorino U, et al: EML4-ALK rearrangement in non-small cell lung
cancer and non-tumor lung tissues. Am J Pathol. 174:661–670. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong DW, Leung EL, So KK, Tam IY, Sihoe
AD, Cheng LC, Ho KK, Au JS, Chung LP, Pik Wong M, et al: The
EML4-ALK fusion gene is involved in various histologic types of
lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer.
115:1723–1733. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takahashi T, Sonobe M, Kobayashi M,
Yoshizawa A, Menju T, Nakayama E, Mino N, Iwakiri S, Sato K,
Miyahara R, et al: Clinicopathologic features of non-small-cell
lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 17:889–897.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang X, Zhang S, Yang X, Yang J, Zhou Q,
Yin L, An S, Lin J, Chen S, Xie Z, et al: Fusion of EML4 and ALK is
associated with development of lung adenocarcinomas lacking EGFR
and KRAS mutations and is correlated with ALK expression. Mol
Cancer. 9:1882010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shaozhang Z, Xiaomei L, Aiping Z, Jianbo
H, Xiangqun S and Qitao Y: Detection of EML4-ALK fusion genes in
non-small cell lung cancer patients with clinical features
associated with EGFR mutations. Genes Chromosomes Cancer.
51:925–932. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Li Y, Yang T, Wei S, Wang J, Wang M,
Wang Y, Zhou Q, Liu H and Chen J: Clinical significance of EML4-ALK
fusion gene and association with EGFR and KRAS gene mutations in
208 Chinese patients with non-small cell lung cancer. PLoS One.
8:e520932013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pomerleau CS, Pomerleau OF, Snedecor SM
and Mehringer AM: Defining a never-smoker: Results from the
nonsmokers survey. Addict Behav. 29:1149–1154. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kwak EL, Bang YJ, Camidge DR, Shaw AT,
Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, et al:
Anaplastic lymphoma kinase inhibition in non-small-cell lung
cancer. N Engl J Med. 363:1693–1703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boland JM, Erdogan S, Vasmatzis G, Yang P,
Tillmans LS, Johnson MR, Wang X, Peterson LM, Halling KC, Oliveira
AM, et al: Anaplastic lymphoma kinase immunoreactivity correlates
with ALK gene rearrangement and transcriptional up-regulation in
non-small cell lung carcinomas. Hum Pathol. 40:1152–1158. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi YL, Takeuchi K, Soda M, Inamura K,
Togashi Y, Hatano S, Enomoto M, Hamada T, Haruta H, Watanabe H, et
al: Identification of novel isoforms of the EML4-ALK transforming
gene in non-small cell lung cancer. Cancer Res. 68:4971–4976. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shaw AT, Yeap BY, Solomon BJ, Riely GJ,
Gainor J, Engelman JA, Shapiro GI, Costa DB, Ou S-HI, Butaney M, et
al: Effect of crizotinib on overall survival in patients with
advanced non-small-cell lung cancer harbouring ALK gene
rearrangement: Aretrospective analysis. Lancet Oncol. 12:1004–1012.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Camidge DR, Bang YJ, Kwak EL, Iafrate AJ,
Varella-Garcia M, Fox SB, Riely GJ, Solomon B, Ou SH, Kim DW, et
al: Activity and safety of crizotinib in patients with ALK-positive
non-small-cell lung cancer: Updated results from a phase 1 study.
Lancet Oncol. 13:1011–1019. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shaw AT, Kim DW, Nakagawa K, Seto T, Crino
L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Solomon BJ, Mok T, Kim D-W, Wu Y-L,
Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et
al: First-line crizotinib versus chemotherapy in ALK-positive lung
cancer. N Engl J Med. 371:2167–2177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Inamura K, Takeuchi K, Togashi Y, Hatano
S, Ninomiya H, Motoi N, Mun MY, Sakao Y, Okumura S, Nakagawa K, et
al: EML4-ALK lung cancers are characterized by rare other
mutations, a TTF-1 cell lineage, an acinar histology and young
onset. Mod Pathol. 22:508–515. 2009. View Article : Google Scholar : PubMed/NCBI
|