Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies and its prognosis is poor with metastasis
(1,2).
HCC cells migrate at an early stage (3). One of the problems associated with HCC
is that its cells migrate into the surrounding fibrous tissues and
proliferate (4). In vitro, HCC
cells can proliferate without nutrients or growth factors such as
fetal bovine serum (FBS) (5). It is
important to detect molecular events at an early stage of HCC cell
migration to improve HCC treatment. Matrix metalloproteinase 9
(MMP9) expression level is upregulated 20-fold after co-culture of
HCC cells and parenchymal cells (6).
MMP9 is a gelatinase involved in cancer metastasis
(7). The overexpression of MMP9 is
associated with the poor prognosis of cancer (8). Research focused on elucidating the
mechanisms involved in the upregulation of MMP9 expression is
therefore important. Polymorphism of the promoter region of MMP9
(−1562)C/T increases the risk of metastasis (9). MMP9 is upregulated in the presence of
inflammatory cytokines such as interleukins 8 and 17 (10). However, details of the mechanisms
involved in the upregulation of MMP9 expression in HCC are not
clear.
The Wnt signaling pathway, which is activated when
Wnt ligands bind their Frizzled (Fz) receptors, has been implicated
in HCC (11). The small interfering
(si)RNA of Fz9 suppresses the proliferation and migration of HCC
cells (12). Studies suggest that the
Wnt pathway is specifically involved in the migration of HCC cells,
and it is expected that inhibition of this pathway may improve the
survival rate of HCC patients by suppressing the proliferation and
migration of HCC cells. One limitation of siRNA, however, is that
its antitumor effect depends on its transfection efficiency
(13).
Niclosamide was originally developed for the
treatment of tapeworm infection and is clinically used worldwide
(14,15). Sack et al first suggested that
niclosamide may be a promising candidate for the treatment of
colorectal cancer through the inhibition of HCC cell proliferation
(16) via the modulation of the Wnt
pathway (17). The mechanisms
proposed for the Wnt pathway-mediated inhibitory effects of
niclosamide include internalization of Fz1 (18) and inhibition of the canonical Wnt
signaling pathway (19). Niclosamide
also downregulates the expression of cyclin D1, one of the target
molecules of the Wnt pathway, via glycogen synthase kinase-3β
(17,20). Thus far, no study has reported the
effects of niclosamide on the migration of HCC cells or on its
regulation of MMP9 expression.
The present study was therefore conducted to
investigate the effects of niclosamide on the migration of HCC
cells and the expression of MMP9.
Materials and methods
Cell culture
HCC HLF and PLC/PRL/5 cell lines were purchased from
Riken Cell Bank (Tsukuba, Japan). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich, St.
Louis, MO, USA) supplemented with 10% FBS (Life Technologies, Grand
Island, NY, USA) and incubated in a humidified chamber at 37°C in
5% carbon dioxide.
Scratch assay
The cells were plated on 4-well chamber slides (BD
Biosciences, Franklin Lakes, NJ, USA) and allowed to yield a
confluent monolayer sheet of cells, which were then scratched with
200-µl pipettes, incubated for 48 h and stained with hematoxylin
and eosin. The stained slides were observed under an AX80
microscope (Olympus, Tokyo, Japan). The distance from the location
of the original scratched line to the new growing edge of the cells
was measured at five different points. The migration distance for
each plate was calculated as the mean average of these five
measurements.
Cell proliferation analysis
The HLF and PRL/PRF/5 cells were trypsinized,
harvested, seeded onto 96-well flat-bottomed plates (Asahi Techno
Glass, Tokyo, Japan) at a density of 1,000 cells per well and then
incubated for 24 h in DMEM supplemented with 10% FBS. Subsequent to
culturing, the cells were treated with niclosamide (Sigma-Aldrich)
at various concentrations (0, 0.01, 0.03, 0.1, 0.3, 1, 3 and 10 µM)
for 72 h and subjected to MTS assay (Promega Corporation, Madison,
WI, USA) according to the manufacturer's instructions. MTS is
bio-reduced by cells into a colored formazan product that can be
detected at a specific wavelength. The absorbance of each reaction
plate was measured at a wavelength of 490 nm using an iMark
Microplate Absorbance Reader (Bio-Rad Laboratories Inc., Hercules,
CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The cells were cultured in 6-well plates (Asahi
Glass Co., Ltd., Tokyo, Japan) and treated with niclosamide for 48
h, after which total RNA (5 µg) was isolated using Isogen (Nippon
Gene Co., Ltd., Tokyo, Japan). The RNA was converted reverse
transcribed into cDNA using the Super Script III and oligo(dT)
primers (Life Technologies Ltd., Carlsbad, CA, USA) according to
the manufacturer's instructions. The PCR primers were as follows:
Ribosomal protein L19 (RPL19; BC095445), forward
5′-CGAATGCCAGAGAAGGTCAC-3′ and reverse 5′-CCATGAGAATCCGCTTGTTT-3′
(157 bp); cyclin D1 (NM_053056), forward 5′-AGAGGCGGAGGAGAACA
AACAG-3′ and reverse 5′-AGGCGGTAGTAGGACAG GAAGTTG-3′ (180 bp); and
MMP-9 (NM_004994) forward 5′-CCTGGGCAGATTCCAAACCT-3′ and reverse
5′-GCAAGTCTTCCGAGTAGTTTTGGAT-3′ (89 bp). RT-qPCR was performed
using Fast SYBR Green Master Mix (Life Technologies Ltd.) for 40
cycles, using 5 sec for denaturation at 95°C and 5 sec for
annealing-extension at 60°C with the Mini Opticon System (Bio Rad,
Hercules, CA). RPL19 was used as an internal control.
Statistical analysis
One-factor analysis of variance was performed on the
data from the cell proliferation studies, RT-qPCR and scratch
assays using JMP10.0.2 software (SAS Institute, Cary, NC, USA).
P<0.05 was used to indicate a statistically significant
difference.
Results
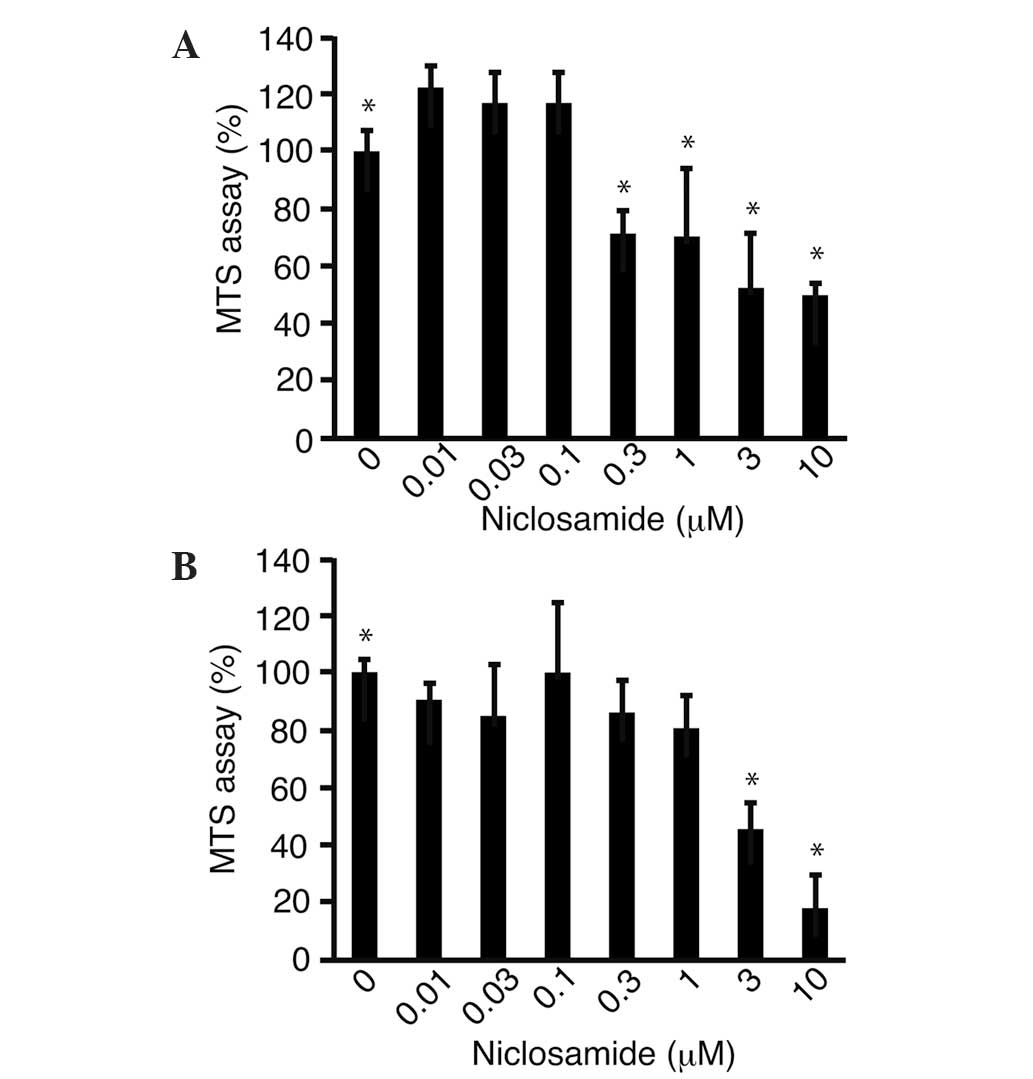
The effects of niclosamide on the proliferation of
the HLF (Fig. 1A) and PRL/PRF/5
(Fig. 1B) cells were examined using
the MTS assay. The proliferation of the HLF and PRL/PRF/5 cells
treated with niclosamide (10 µm) was significantly (P<0.05)
suppressed to 49.9±3.7 and 17.9±11.5%, respectively, compared with
that of the untreated control cells.
Cyclin D1 is a downstream molecule associated with
the Wnt pathway and involved in cell proliferation (21). The HLF and PRL/PRF/5 cells were
cultured with niclosamide. The expression levels of cyclin D1 in
the HLF (Fig. 2A) and PRL/PRF/5
(Fig. 2B) cells cultured in the
presence of niclosamide were analyzed using RT-qPCR. The results
showed that cyclin D1 expression was significantly (P<0.05)
downregulated to 52.4±4.4 and 23.9±5.4% in the HLF and PRL/PRF/5
cells, respectively, compared with the levels in the untreated
control cells.
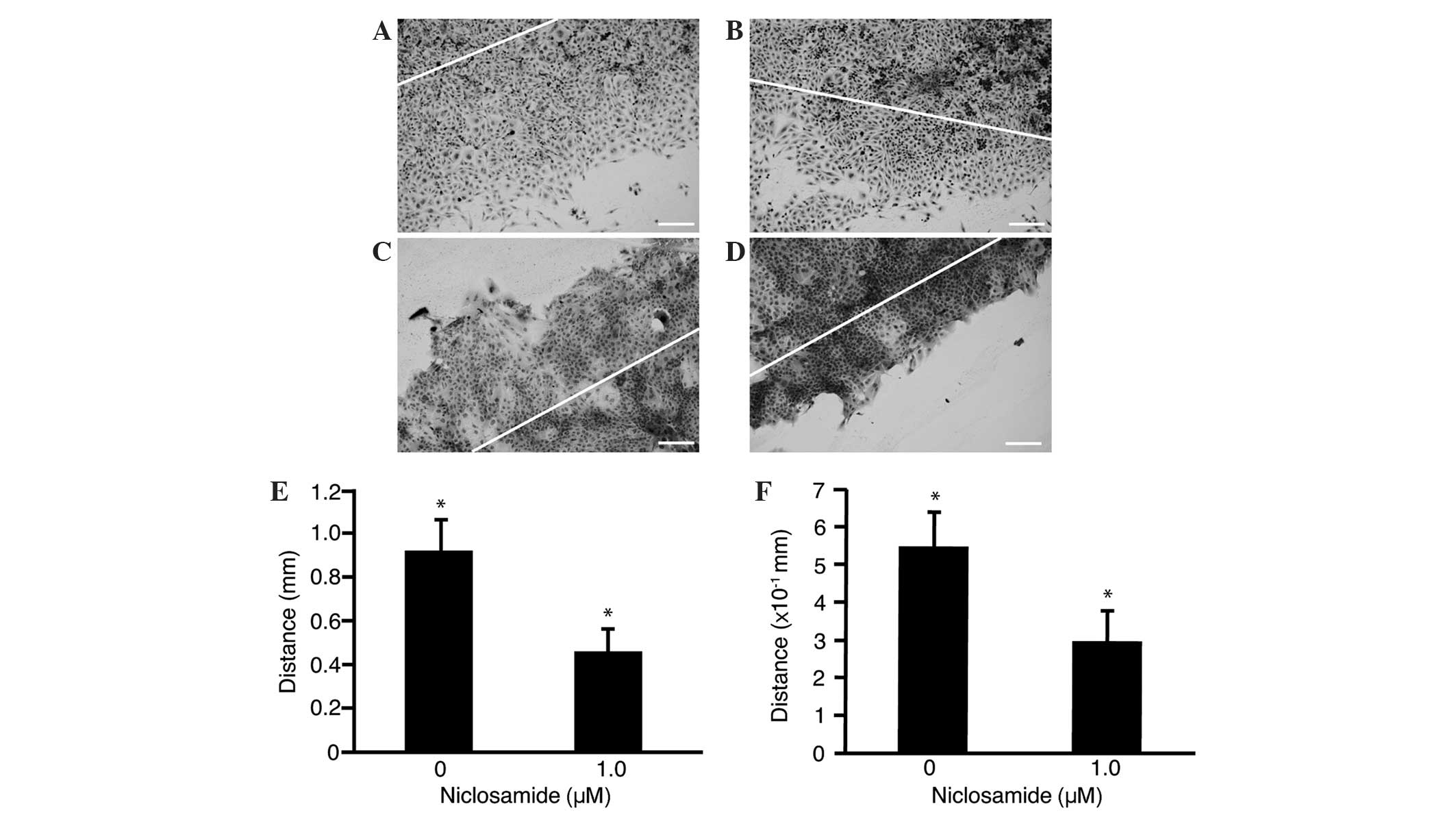
Change in cell motility was also investigated in the
HLF (Fig. 3A and B) and PRL/PRF/5
(Fig. 3C and D) cells treated with
niclosamide (1.0 µm). A reduction in the distance of the scratched
line from the growing edge was observed in the treated cells
(Fig. 3B and D) compared with the
untreated control cells (Fig. 3A and
C). The HLF (Fig. 3E) and
PRL/PRF/5 (Fig. 3F) cells treated
with niclosamide showed significant (P<0.05) decreases in cell
migration (4.6±1.0 and 3.0±0.8 mm, respectively) compared with that
of the untreated control HLF and PRL/PRF/5 cells (9.2±1.4 and
5.5±0.9 mm, respectively).
MMP9 is involved in cell motility, and as expected,
its expression level in the HLF (Fig.
4A) and PRL/PRF/5 (Fig. 4B) cells
treated with niclosamide (1.0 µm) was significantly (P<0.05)
decreased to 22.4±1.76% and 18.7±10.7% compared with that in the
untreated cells.
Discussion
Niclosamide is currently used to treat tapeworm
infection (15). Application of
niclosamide in the treatment of cancer was first reported for
colorectal cancer (16). Niclosamide
suppresses the proliferation of HCC cells by inhibiting the Wnt
signaling pathway (17).
Niclosamide has been previously shown to suppress
the migration of breast cancer cells and colon cancer cells
(16,22). The present results revealed a similar
suppression of HCC cell migration by niclosamide, suggesting that
it may suppress the migration of cancer cells. These results are
promising and may be clinically significant, as niclosamide could
improve the prognosis of cancer patients by suppressing metastasis.
While the mechanism by which niclosamide suppresses the migration
of cancer cells was not completely clear, the present study clearly
showed that MMP9 expression was downregulated by niclosamide,
thereby strongly suggesting its involvement in the suppression of
cancer cell migration. Previous studies and the present study have
indicated that niclosamide is promising for the treatment of HCC
(23).
The present data clearly showed that niclosamide
suppressed the migration of HCC cells, and that MMP9 decreased with
niclosamide treatment. This suggests that niclosamide suppresses
migration via inhibiting the expression of MMP9 (7). However, the mechanism underlying
decreased MMP9 expression remains unclear. Our previous study
revealed that niclosamide suppresses the Wnt pathway (17). Therefore, it is suggested that the
expression of MMP9 decreases with suppression of the Wnt pathway,
and that MMP9 may be a downstream molecule of the Wnt pathway.
In conclusion, niclosamide significantly suppressed
the migration of the HCC cells, probably by mechanisms involving
the downregulation of MMP9 expression. Future studies in line with
these results could involve investigation of the promoter activity
of MMP9, which would further elucidate the effects of niclosamide
at a transcriptional level.
References
|
1
|
Tabrizian P, Roayaie S and Schwartz ME:
Current management of hepatocellular carcinoma. World J
Gastroenterol. 20:10223–10237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang H and Chen L: Tumor microenviroment
and hepatocellular carcinoma metastasis. J Gastroenterol Hepatol.
28(Suppl 1): S43–S48. 2013. View Article : Google Scholar
|
|
3
|
Kondo F, Kondo Y, Nagato Y, Tomizawa M and
Wada K: Interstitial tumour cell invasion in small hepatocellular
carcinoma. Evaluation in microscopic and low magnification views. J
Gastroenterol Hepatol. 9:604–612. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tomizawa M, Kondo F and Kondo Y: Growth
patterns and interstitial invasion of small hepatocellular
carcinoma. Pathol Int. 45:352–358. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tomizawa M, Shinozaki F, Sugiyama T,
Yamamoto S, Sueishi M and Yoshida T: Sorafenib suppresses the cell
cycle and induces the apoptosis of hepatocellular carcinoma cell
lines in serum-free media. Exp Ther Med. 1:863–866. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen RX, Song HY, Dong YY, Hu C, Zheng QD,
Xue TC, Liu XH, Zhang Y, Chen J, Ren ZG, et al: Dynamic expression
patterns of differential proteins during early invasion of
hepatocellular carcinoma. PLoS One. 9:e885432014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vandooren J, Van den Steen PE and
Opdenakker G: Biochemistry and molecular biology of gelatinase B or
matrix metalloproteinase-9 (MMP-9): The next decade. Crit Rev
Biochem Mol Biol. 48:222–272. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li H, Zhang K, Liu LH, Ouyang Y, Bu J, Guo
HB and Xiao T: A systematic review of matrix metalloproteinase 9 as
a biomarker of survival in patients with osteosarcoma. Tumour Biol.
35:5487–5491. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu D, Guo H, Li Y, Xu X, Yang K and Bai
Y: Association between polymorphisms in the promoter regions of
matrix metalloproteinases (MMPs) and risk of cancer metastasis: A
meta-analysis. PLoS One. 7:e312512012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zarogoulidis P, Katsikogianni F, Tsiouda
T, Sakkas A, Katsikogiannis N and Zarogoulidis K: Interleukin-8 and
interleukin-17 for cancer. Cancer Invest. 32:197–205. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pez F, Lopez A, Kim M, Wands JR, de Caron
Fromentel C and Merle P: Wnt signaling and hepatocarcinogenesis:
Molecular targets for the development of innovative anticancer
drugs. J Hepatol. 59:1107–1117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujimoto T, Tomizawa M and Yokosuka O:
SiRNA of frizzled-9 suppresses proliferation and motility of
hepatoma cells. Int J Oncol. 35:861–866. 2009.PubMed/NCBI
|
|
13
|
Wang YQ, Wang F, Deng XQ, Sheng J, Chen SY
and Su J: Delivery of therapeutic AGT shRNA by PEG-Bu for
hypertension therapy. PLoS One. 8:e686512013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Knorr R: Treatment of tapeworm with
Yomesan in 36 patients. Med Lav. 55:1937–1938. 1960.(In German).
PubMed/NCBI
|
|
15
|
Mwape KE, Phiri IK, Praet N, Muma JB, Zulu
G, Van den Bossche P, de Deken R, Speybroeck N, Dorny P and Gabriël
S: Taenia solium Infections in a rural area of Eastern Zambia-a
community based study. PLoS Negl Trop Dis. 6:e15942012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sack U, Walther W, Scudiero D, Selby M,
Kobelt D, Lemm M, Fichtner I, Schlag PM, Shoemaker RH and Stein U:
Novel effect of antihelminthic Niclosamide on S100A4-mediated
metastatic progression in colon cancer. J Natl Cancer Inst.
103:1018–1036. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tomizawa M, Shinozaki F, Motoyoshi Y,
Sugiyama T, Yamamoto S, Sueishi M and Yoshida T: Niclosamide
suppresses Hepatoma cell proliferation via the Wnt pathway. Onco
Targets Ther. 6:1685–1693. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen M, Wang J, Lu J, Bond MC, Ren XR,
Lyerly HK, Barak LS and Chen W: The anti-helminthic niclosamide
inhibits Wnt/Frizzled1 signaling. Biochemistry. 48:10267–10274.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wieland A, Trageser D, Gogolok S, Reinartz
R, Höfer H, Keller M, Leinhaas A, Schelle R, Normann S, Klaas L, et
al: Anticancer effects of niclosamide in human glioblastoma. Clin
Cancer Res. 19:4124–4136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takahashi-Yanaga F and Sasaguri T:
GSK-3beta regulates cyclin D1 expression: A new target for
chemotherapy. Cell Signal. 20:581–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takayasu H, Horie H, Hiyama E, Matsunaga
T, Hayashi Y, Watanabe Y, Suita S, Kaneko M, Sasaki F, Hashizume K,
et al: Frequent deletions and mutations of the beta-catenin gene
are associated with overexpression of cyclin D1 and fibronectin and
poorly differentiated histology in childhood hepatoblastoma. Clin
Cancer Res. 7:901–908. 2001.PubMed/NCBI
|
|
22
|
Ye T, Xiong Y, Yan Y, Xia Y, Song X, Liu
L, Li D, Wang N, Zhang L, Zhu Y, et al: The anthelmintic drug
niclosamide induces apoptosis, impairs metastasis and reduces
immunosuppressive cells in breast cancer model. PLoS One.
9:e858872014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Li PK, Roberts MJ, Arend RC, Samant
RS and Buchsbaum DJ: Multi-targeted therapy of cancer by
niclosamide: A new application for an old drug. Cancer Lett.
349:8–14. 2014. View Article : Google Scholar : PubMed/NCBI
|