Introduction
Extranodal natural killer (NK)/T-cell lymphoma,
nasal type (ENKTCL) is an aggressive type of lymphoma that occurs
frequently in the Asian population (1,2). The
pathogenesis of this tumor is poorly understood, and conventional
chemotherapy regimens, currently employed for the treatment of
other aggressive types of lymphoma, usually provide poor outcomes
in patients with ENKTCL (1,2). Therefore, studies on the pathogenetic
abnormalities that occur during the development of ENKTCL may
contribute to improving the clinical outcomes of these
patients.
MicroRNAs (miRNAs) are small non-coding RNA
molecules that inhibit the transcription or translation of mRNA.
Previous studies have demonstrated that dysregulation of miRNA
occurs in numerous types of human cancer, indicating that miRNAs
may act as oncogenes or tumor suppressor genes (3). Previous miRNA expression profiling
studies conducted on a series of ENKTCL formalin-fixed
paraffin-embedded (FFPE) tissues revealed that several miRNAs,
including miR-10a, miR-22, miR-340, miR-342-3p and miR-590-5p, are
dysregulated in ENKTCL tissues, compared with normal NK cells
(1). Bioinformatic analysis of these
miRNAs indicated that all of them target the T-lymphoma invasion
and metastasis-inducing factor 1 (TIAM1) gene (1).
Tiam1 is a specific guanine nucleotide exchange
factor for Rho-like guanosine triphosphate (GTP)ases, which
exhibits its pathophysiological role via the activation of the rat
sarcoma-related C3 botulinum toxin substrate signaling pathway
(4). Overexpression of TIAM1 has been
reported in various solid tumors (5–12). In
addition, previous studies have demonstrated that TIAM1 modulates a
number of cellular processes considered to be associated with tumor
progression, including cell apoptosis, invasion and migration
(13–18). These findings suggest that TIAM1 may
be a target for cancer therapy. However, there is limited evidence
regarding the role of the TIAM1 gene in the pathogenesis of
ENKTCL.
To gain insight into the potential role of miR-10a,
miR-22, miR-340, miR-342-3p, miR-590-5p and TIAM1 in the
pathogenesis of ENKTCL, the present study examined the expression
levels of these miRNAs and their target gene TIAM1 in ENKTCL
tissues, in order to assess whether the expression levels of these
molecules correlated with the clinical features of patients with
ENKTCL.
Materials and methods
Patients and controls
Patients who were diagnosed with ENKTCL from 2007 to
2011 were selected from the archives of the Department of Pathology
of the Fujian Medical University Union Hospital (Fuzhou, China),
and classified according to the 2008 World Health Organization
classification of lymphomas (19). A
total of 21 patients were selected for the study. The study was
approved by the ethics committee of Fujian Medical University Union
Hospital. The samples were collected with the patients' consent.
Patients with no additional tissue available for
immunohistochemical testing were excluded from the study. The FFPE
tissues of 15 patients were subjected to quantitative polymerase
chain reaction (qPCR) analysis. In addition, ten samples of normal
and reactive lymph node hyperplasia FFPE tissue were included as
controls.
Isolation of normal NK cells from
peripheral blood
Human normal NK cells were isolated from whole blood
samples obtained from healthy donors, which were collected with
EDTA using NK Cell Isolation Kit (TBD Science, Tianjin, China).
RNA extraction
Prior to RNA extraction, five pieces of the FFPE
tissue sections were treated with 1 ml xylene (Kemiou Chemical
Reagent Co., Tianjin, China) for 10 sec at room temperature, then
incubated at 56 degree for 3 min to remove the paraffin, and
subsequently digested with 10 µl proteinase K (Qiagen, Inc.,
Valencia, CA, USA), followed by incubation at 56°C for 15 min, then
at 80°C for 15 min. Total RNA was then isolated using miRNeasy FFPE
Kit (Qiagen, Inc.), according to the manufacturers protocol. Total
RNA from NK cells was extracted with TRIzol (Invitrogen, Thermo
Fisher Scientific, Inc., Waltham, MA, USA).
Reverse transcription (RT) and qPCR of
miRNAs
Complementary DNA was synthesized from total RNA
using PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara Bio,
Inc., Otsu, Japan), following the manufacturers protocol. The RT
reactions contained 500 ng total RNA extracted from the samples, 2
µl 5X PrimeScript™ Buffer (Takara Bio, Inc.), 0.5 µl 1X
PrimeScript™ RT Enzyme Mix I (Takara Bio, Inc.) and 0.5 µl
oligo(dT) primer (Takara Bio, Inc.). The 10-µl reactions were
incubated for 42 min at 37°C, followed by 30-sec incubation at
85°C, and then exposed to 4°C.
To quantify the expression levels of the
aforementioned miRNAs, qPCR was conducted using SYBR® Premix Ex
Taq™ II (Tli RNase H Plus) Kit (Takara, Bio, Inc.) with an Applied
Biosystems 7300 Real Time PCR System (Thermo Fisher Scientific,
Inc.), according to the manufacturers protocol. The 20-µl PCR
reactions included 0.19 µl RT product, 10.0 µl 2X SYBR® Premix Ex
Taq™ II (Takara Bio, Inc.), 2.0 µl primers mix (Biosune, Inc.,
Shanghai, China), 0.4 µl 50X ROX Reference Dye II (Takara Bio,
Inc.) and 7.41 µl RNase-free dH2O (Takara Bio, Inc.).
The reaction mixtures were incubated in a 96-well plate at 95°C for
1 min, followed by 40 cycles of amplification at 95°C for 5 sec and
60°C for 30 sec. The sequences of the primers used are listed in
Table I. The quantification cycle
(Cq) was determined using default threshold settings.
All the experiments were performed in triplicate. U6 small nuclear
RNA was used as control to normalize the miRNA input in the qPCR
assay. The qPCR data were analyzed using the 2−ΔCq
method.
 | Table I.Primer sequences of miRNAs for
quantitative polymerase chain reaction analysis. |
Table I.
Primer sequences of miRNAs for
quantitative polymerase chain reaction analysis.
|
| Primer sequence
(5′-3) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| miRNA-10a | TAC CCT GTA GAT CCG
AAT TTGTG | ATT CCC CTA GAT ACG
AAT TTG TGA |
| miRNA-22 |
AGCAACATGCCCTGCTC |
TCTGTCACCTTCCAGATGATG |
| miRNA-340 |
ATAAAGCAATGAGACTGATTGTC |
GGCTATAAAGTAACTGAGACGGA |
| miRNA-342-3p |
GTGCTATCTGTGATTGAGGGA |
CGGGTGCGATTTCTGTG |
| miRNA-590-5p |
TTAGAGCCAACCAGCAGC |
GCATTGACAGCACATCCC |
| U6 |
GTTTTGTAGTTTTTGGAGTTAGTGTTGTGT |
CTCAACCTACAATCAAAAACAACACAAACA |
Immunohistochemistry
Tissue sections were subjected to antigen retrieval
by incubation in 10 mmol/l sodium citrate buffer (pH 6.0) for 10
min in a microwave oven (WD900SL23–2, Galanz Enterprises Co., Ltd.,
Foshan, China) at the maximum power setting. Any potential
endogenous peroxidase activity present in the tissues was blocked
by incubation with 3% H2O2 for 15 min.
Subsequently, the tissue sections were incubated at room
temperature for 60 min with a rabbit polyclonal antibody specific
for human TIAM1 (dilution,7 1:100; cat no. sc-872, Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) diluted in phosphate-buffered
saline (PBS). Immunoreactive proteins were visualized with
MaxVision™ HRP-Polymer anti-Rabbit IHC Kit (Fuzhou Maixin Biotech
Co., Ltd., Fuzhou, China), following the manufacturers protocol,
and counterstained with hematoxylin (Sigma-Aldrich Shanghai Trading
Co., Ltd., Shanghai, China). Tissue sections corresponding to the
negative control were treated with PBS under the same experimental
conditions than the samples.
Quantitative evaluation of the protein expression
levels of TIAM1 was performed by counting the percentage of
immunoreactive cells positive for TIAM1 that were present in a
number of high-power microscopic fields (magnification, ×200; BX41
microscope, Olympus Corporation, Tokyo, Japan). The ENKTCL tissues
were scored based on the percentage of positive tumor cells
expressing cytoplasmic TIAM1, as follows: i) <15%, score 0; ii)
15–24%, score 1; iii) 25–49%, score 2; iv) 50–74%, score 3; and v)
≥75%, score 4.
Statistical analysis
Since the data corresponding to the expression
levels of miR-10a, miR-22, miR-340, miR-342-3p and miR-590-5p
followed a normal distribution, the expression levels of these
miRNAs in ENKTCL tissues and normal NK cells were compared using
Students two-tailed t test. The association between the clinical
features of patients with ENKTCL and the expression levels of
miR-10a, miR-342-3p and TIAM1 protein detected in these patients
was analyzed via Students two-tailed t test and χ2 test,
respectively. Spearmans rank correlation was used to evaluate the
association between the expression levels of miR-10a and miR-342-3p
and the protein expression levels of TIAM1. P<0.05 was
considered to indicate a statistical significant difference. SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA) was used for
statistical analysis.
Results
Clinical features of patients with
ENKTCL
The main demographic and clinical features of
patients with ENKTCL are listed in Table
II. The median age at the time of diagnosis was 44.1 years
(range, 13–71 years), and the most frequent cancer site was the
nasal cavity. According to the Ann Arbor staging of lymphoma, 6
patients were in stage I, 3 in stage II, 2 in stage III and 10 in
stage IV (20). High expression
levels of lactate dehydrogenase (LDH) were observed in 10 patients,
and 12 patients exhibited B symptoms.
 | Table II.Clinical features of 21 patients with
ENKTCL. |
Table II.
Clinical features of 21 patients with
ENKTCL.
| Patient no. | Age (years) | Gender | Primary cancer
site | Stagea | LDH levels | B symptoms | IPI | TIAM1 in ENKTCL
(IHS)b |
|---|
| 1 | 27 | Male | Nasal cavity | I | Normal | A | 0 | 0 |
| 2 | 52 | Female | Nasal cavity | II | Normal | B | 0 | 0 |
| 3 | 71 | Male | Nasal cavity | I | Normal | A | 1 | 0 |
| 4 | 68 | Male | Gingiva | IV | Normal | A | 4 | 0 |
| 5 | 24 | Male | Adrenal gland | III | Normal | A | 2 | 2 |
| 6 | 54 | Male | Nasal cavity | IV | High | B | 4 | 0 |
| 7 | 55 | Male | Nasal cavity | I | Normal | A | 0 | 2 |
| 8 | 45 | Female | Bone marrow | IV | Normal | B | 3 | 2 |
| 9 | 39 | Male | Nasal cavity | IV | High | B | 3 | 1 |
| 10 | 41 | Male | Nasal cavity | I | High | A | 1 | 2 |
| 11 | 26 | Male | Gastric | IV | High | A | 4 | 0 |
| 12 | 64 | Male | Nasal cavity | I | High | A | 3 | 1 |
| 13 | 13 | Female | Bone marrow | IV | High | B | 4 | 2 |
| 14 | 55 | Male | Bone marrow | IV | High | B | 4 | 1 |
| 15 | 43 | Male | Bone marrow | IV | High | B | 4 | 0 |
| 16 | 43 | Female | Nasal cavity | II | High | B | 2 | 0 |
| 17 | 71 | Male | Nasal cavity | I | Normal | A | 2 | 0 |
| 18 | 40 | Male | Nasal cavity | IV | Normal | B | 3 | 1 |
| 19 | 39 | Male | Nasal cavity | III | Normal | B | 3 | 2 |
| 20 | 24 | Male | Nasal cavity | IV | High | B | 3 | 2 |
| 21 | 32 | Male | Nasal cavity | II | Normal | B | 1 | 0 |
qPCR results of miR-10a, miR-22,
miR-340, miR-342-3p and miR-590-5p
The expression levels of miR-10a were markedly lower
in ENKTCL tissues than in normal NK cells (Fig. 1A). In addition, the expression levels
of miR-342-3p in ENKTCL tissues were significantly lower than in
normal NK cells (Fig. 1B). In
contrast, the expression levels of miR-22, miR-340 and miR-590-5p
did not differ significantly between ENKTCL tissues and normal NK
cells (Fig. 1C–E). These results
suggest that miR-10a and miR-342-3p may be involved in the
pathogenesis of ENKTCL.
Correlations between the expression
levels of miR-10a and miR-342-3p and the demographic and clinical
characteristics of patients with ENKTCL
The expression levels of miR-10a in ENKTCL FFPE
tissues were inversely correlated with the patients age (P=0.02),
but not with other demographic or clinical features, including
gender, Ann Arbor stage, levels of LDH, B symptoms and
international prognostic index (IPI) score (Table III). The expression levels of
miR-342-3p in ENKTCL FFPE tissues was not correlated with any
demographic or clinical features of the patients.
 | Table III.Association between the expression
levels of miR-10a, miR-342-3p and TIAM1 protein in tissues of
patients with ENKTCL and the demographic and clinical features of
the patients. |
Table III.
Association between the expression
levels of miR-10a, miR-342-3p and TIAM1 protein in tissues of
patients with ENKTCL and the demographic and clinical features of
the patients.
|
|
| miR-10a | miR-342-3p | TIAM1 |
|---|
|
|
|
|
|
|
|---|
| Clinical
feature | Cases (no.) | Expression
(×10−3)a |
P-valueb | Expression
(×10−3)a |
P-valueb | Cases (no.) | TIAM1+
cases (no.) |
P-valuec |
|---|
| Age (years) |
|
| 0.02 |
| 0.97 |
|
| 0.34 |
|
﹤60 | 13 | 4.97±1.13 |
| 2.08±0.27 |
| 17 | 10 |
|
|
≥60 | 2 | 1.46±0.64 |
| 2.05±0.73 |
| 4 | 1 |
|
| Gender |
|
| 0.58 |
| 0.70 |
|
| 0.92 |
|
Male | 12 | 4.03±1.02 |
| 2.02±0.28 |
| 17 | 9 |
|
|
Female | 3 | 6.41±3.43 |
| 2.29±0.56 |
| 4 | 2 |
|
| Ann Arbor
stage |
|
| 0.98 |
| 0.22 |
|
| 0.02 |
|
I–II | 6 | 4.46±1.98 |
| 1.68±0.41 |
| 9 | 2 |
|
|
III–IV | 9 | 4.54±1.20 |
| 2.34±0.28 |
| 12 | 9 |
|
| LDH levels |
|
| 0.93 |
| 0.90 |
|
| 0.51 |
|
High | 6 | 4.39±1.72 |
| 2.12±0.44 |
| 10 | 6 |
|
|
Normal | 9 | 4.59±1.36 |
| 2.05±0.3 |
| 11 | 5 |
|
| B symptoms |
|
| 0.06 |
| 0.88 |
|
| 0.53 |
|
Positive | 8 | 6.28±1.53 |
| 2.11±0.32 |
| 12 | 7 |
|
|
Negative | 7 | 2.48±0.93 |
| 2.04±0.39 |
| 9 | 4 |
|
| IPI (score) |
|
| 0.61 |
| 0.21 |
|
| 0.02 |
|
0–2 | 7 | 3.90±1.77 |
| 1.74±0.35 |
| 9 | 2 |
|
|
3–5 | 8 | 5.04±1.24 |
| 2.37±0.32 |
| 12 | 9 |
|
Immunohistochemistry and correlation
between the expression levels of TIAM1 protein and the demographic
and clinical features of patients with ENKTCL
Tiam1 protein was expressed in 11 ENKTCL samples
(52.4%) and in 1 of 10 paired samples of normal and reactive lymph
node hyperplasia (10%), where its expression levels were low
(Fig. 2). The intensity of TIAM1
protein expression detected in the ENKTCL tissues is listed in
Table II. The protein expression
levels of TIAM1 in ENKTCL FFPE tissues were positively correlated
with Ann Arbor stage and IPI score (P=0.02; Table III), but no significant association
was observed with any other demographic or clinical features of the
patients. These results suggest that TIAM1 protein may be involved
in the pathogenesis of ENKTCL.
Correlations between the expression
levels of miR-10a, miR-342-3p and TIAM1 protein in ENKTCL FFPE
tissues
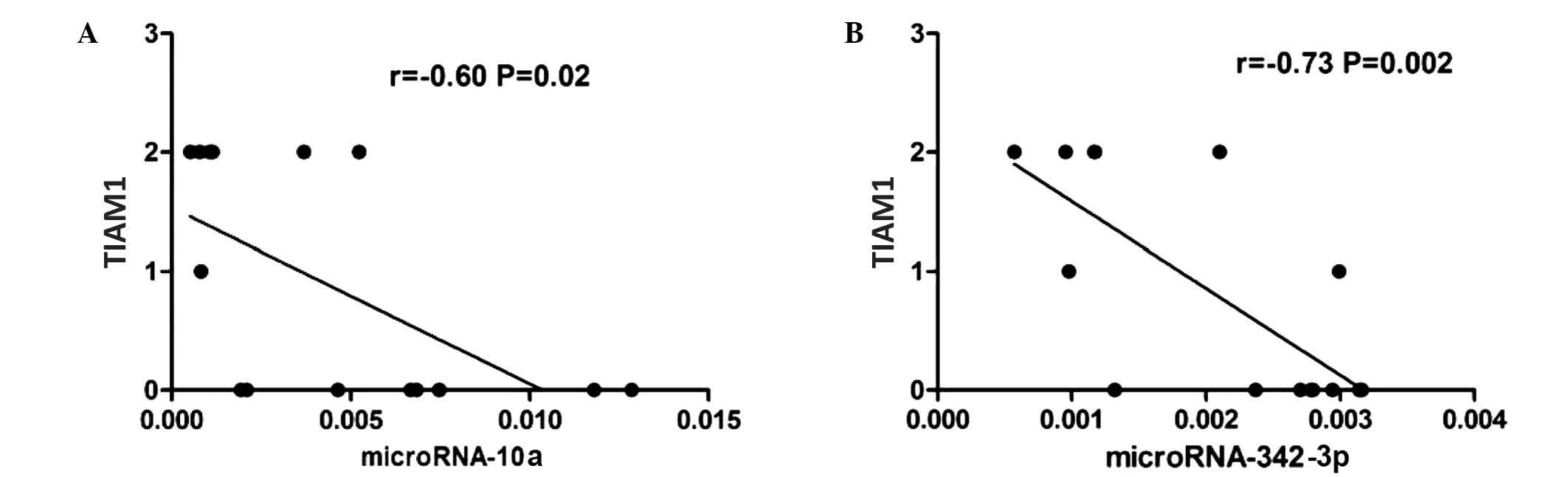
Expression of TIAM1 protein was detected in the FFPE
tissues of 7 of the 15 patients with ENKTCL analyzed (47%). In
these patients, the expression levels of miR-10a and miR-342-3p
appeared to be inversely correlated with the protein expression
levels of TIAM1 (Spearmans r=−0.60 and −0.73 for miR-10a and
miR-342-3p, respectively; P=0.02; Fig.
3).
Discussion
Previous studies have demonstrated that the
expression levels of several miRNAs are downregulated in ENKTCL
FFPE tissues, which is considered to contribute to the pathogenesis
of the tumor by the loss of the suppressive effects that these
miRNAs normally exert on their target genes (1,21–25). In a study by Ng et al (1), the expression levels of miR-10a, miR-22,
miR-340, miR-342-3p and miR-590-5p appeared to be downregulated in
ENKTCL FFPE tissues, compared with normal NK cells, according to
the results of human miRNA microarray analysis. However, these
findings were not further validated by qPCR.
The qPCR data of the present study are consistent
with the previous miRNA microarray results reported by Ng et
al (1), confirming that miR-10a
and miR-342-3p are downregulated in ENKTCL FFPE tissues, compared
with normal NK cells. However, in the present study, the expression
levels of miR-22, miR-340 and miR-590-5p did not differ
significantly between ENKTCL tissues and normal NK cells.
Furthermore, the expression levels of miR-10a in the ENKTCL FFPE
tissues correlated with the patients age, but the expression levels
of miR-342-3p did not correlate with any demographic or clinical
feature of the patients. Therefore, further studies are required to
validate the potential participation of miR-10a and miR-342-3p in
the pathogenesis of ENKTCL.
Previous studies have identified several target
genes of miR-10a, including high-mobility group A2 (26), cell adhesion molecule L1-like
(27), homeobox (HOX)A1 (28) and HOXD4 (29), which are involved in cellular
differentiation, growth, migration and invasion in various
pathophysiological processes (26–29). Other
studies have demonstrated that the expression of miR-342-3p is
downregulated in the blood and tumor tissues of patients with
cancer, including colorectal cancer (30), clinical glioblastoma multiforme
(31), breast tumor (32), acute lymphoblastic leukemia (33) and Sézary syndrome (34). In addition, miR-342-3p has been
previously demonstrated to be involved in cell differentiation
(35), growth (36,37),
invasion (37) and response to
chemotherapy (38,39) in cancer cells.
Previous studies have suggested that multiple miRNAs
may suppress the expression of the same target gene by directly
targeting its 3 untranslated region (40). Furthermore, previous bioinformatic
analysis indicated that miR-10a and miRNA-342-3p target the TIAM1
gene (1).
Tiam1 has been identified as a guanine nucleotide
exchange factor that exchanges guanosine diphosphate for GTP in
Rho-like GTPases, thereby activating them. This process leads to
the activation of a signaling pathway that stimulates the c-Jun
N-terminal kinase, p38 mitogen-activated protein kinase and
extracellular signal-regulated kinase pathways, which results in
the regulation of the expression of genes involved in cellular
migration, invasion and metastasis (4,5). To date,
overexpression of TIAM1 has been reported in various types of tumor
tissue, including head and neck (5),
esophageal (6), colorectal (7), gallbladder (8), renal cell (9), nasopharyngeal (10), hepatocellular (11) and prostate carcinoma (12). In addition, overexpression of TIAM1
has been suggested to be involved in tumor progression via
lymphangiogenesis (13), apoptosis
(14,15), invasion and migration (16–18).
However, the expression of TIAM1 in ENKTCL FFPE tissue and its
association with clinical features of patients with ENKTCL remain
unclear.
The immunohistochemistry data of the present study
demonstrated that TIAM1 protein was overexpressed in ENKTCL FFPE
tissues, compared with normal and reactive lymph node hyperplasia
FFPE tissues, which is in agreement with the results of previous
studies on various malignancies (4–6,8–13). The overexpression of
TIAM1 in the ENKTCL cases analyzed in the present study correlated
with the Ann Arbor stage and IPI score of the tumors. However, no
significant association was observed between the protein expression
levels of TIAM1 and any other demographic and clinical
characteristics of the patients, including age, gender, levels of
LDH and B symptoms. Overall, the results of the present study
suggest that TIAM1 may be involved in the pathogenesis of ENKTCL.
However, due to the small sample size of the present study, further
studies involving a larger number of cases of ENKTCL are required
in order to confirm these findings.
In conclusion, the results of the present study
suggest that reduced expression of miR-10a and miR-342-3p and
overexpression of TIAM1 protein may be involved in the progression
of ENKTCL. Additional in vitro and in vivo studies
are required to further elucidate the potential role and mechanism
of action of these molecules in the development of ENKTCL.
Acknowledgements
The present study was funded by the National Natural
Science Foundation of China (Beijing, China; grant no. 81201872),
the Natural Science Foundation of Fujian Province (Fuzhou, China;
grant no. 2013J01308), the Fujian Provincial Health Bureau Youth
Research Projects (Fuzhou, China; grant no. 2011-1-12), the Medical
Elite Cultivation Program of Fujian Province (Fuzhou, China; grant
no. 2015-ZQN-JC-18) and the Middle Age and Young Teacher Education
and Research Program of Fujian Province (Fuzhou, China; grant no.
JA14133).
References
|
1
|
Ng SB, Yan J, Huang G, Selvarajan V, Tay
JL, Lin B, Bi C, Tan J, Kwong YL, Shimizu N, et al: Dysregulated
microRNAs affect pathways and targets of biologic relevance in
nasal-type natural killer/T-cell lymphoma. Blood. 118:4919–4929.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim SJ, Jung HA, Chuang SS, Hong H, Guo
CC, Cao J, Hong XN, Suzuki R, Kang HJ, Won JH, et al: Asia Lymphoma
Study Group: Extranodal natural killer/T-cell lymphoma involving
the gastrointestinal tract: Analysis of clinical features and
outcomes from the Asia Lymphoma Study Group. J Hematol Oncol.
6:862013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cook DR, Rossman KL and Der CJ: Rho
guanine nucleotide exchange factors: Regulators of Rho GTPase
activity in development and disease. Oncogene. 33:4021–4035. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang S, Li S, Yang X, Yang S, Liu S, Liu B
and Liu J: Elevated expression of T-lymphoma invasion and
metastasis inducing factor 1 in squamous-cell carcinoma of the head
and neck and its clinical significance. Eur J Cancer. 50:379–387.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu H, Shi G, Liu X, Wu H, Fan Q and Wang
X: Overexpression of Tiam1 predicts poor prognosis in patients with
esophageal squamous cell carcinoma. Oncol Rep. 25:841–848.
2011.PubMed/NCBI
|
|
7
|
Jin H, Li T and Ding Y, Deng Y, Zhang W,
Yang H, Zhou J, Liu C, Lin J and Ding Y: Methylation status of
T-lymphoma invasion and metastasis 1 promoter and its
overexpression in colorectal cancer. Hum Pathol. 42:541–551. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du X, Wang S, Lu J, Wang Q, Song N, Yang
T, Dong R, Zang L, Yang Y, Wu T and Wang C: Clinical value of
Tiam1-Rac1 signaling in primary gallbladder carcinoma. Med Oncol.
29:1873–1878. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao L, Liu Y, Sun X, He M and Ding Y:
Overexpression of T lymphoma invasion and metastasis 1 predict
renal cell carcinoma metastasis and overall patient survival. J
Cancer Res Clin Oncol. 137:393–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qi Y, Huang B, Yu L, Wang Q, Lan G and
Zhang Q: Prognostic value of Tiam1 and Rac1 overexpression in
nasopharyngeal carcinoma. ORL J Otorhinolaryngol Relat Spec.
71:163–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding Y, Chen B, Wang S, Zhao L, Chen J,
Ding Y, Chen L and Luo R: Overexpression of Tiam1 in hepatocellular
carcinomas predicts poor prognosis of HCC patients. Int J Cancer.
124:653–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Engers R, Mueller M, Walter A, Collard JG,
Willers R and Gabbert HE: Prognostic relevance of Tiam1 protein
expression in prostate carcinomas. Br J Cancer. 95:1081–1086. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhong D, Li Y, Peng Q, Zhou J, Zhou Q,
Zhang R and Liang H: Expression of Tiam1 and VEGF-C correlates with
lymphangiogenesis in human colorectal carcinoma. Cancer Biol Ther.
8:689–695. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao-Hong, Shibayama-Imazu T, Masuda Y,
Shinki T, Nakajo S and Nakaya K: Involvement of Tiam1 in apoptosis
induced by bufalin in HeLa cells. Anticancer Res. 27:245–249.
2007.PubMed/NCBI
|
|
15
|
Minard ME, Ellis LM and Gallick GE: Tiam1
regulates cell adhesion, migration and apoptosis in colon tumor
cells. Clin Exp Metastasis. 23:301–313. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Adam L, Vadlamudi RK, McCrea P and Kumar
R: Tiam1 overexpression potentiates heregulin-induced lymphoid
enhancer factor-1/β-catenin nuclear signaling in breast cancer
cells by modulating the intercellular stability. J Biol Chem.
276:28443–28450. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Engers R, Springer E, Michiels F, Collard
JG and Gabbert HE: Rac affects invasion of human renal cell
carcinomas by upregulating tissue inhibitor of metalloproteinases
(TIMP)-1 and TIMP-2 expression. J Biol Chem. 276:41889–41897. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bourguignon LY, Zhu H, Shao L and Chen YW:
Ankyrin-Tiam1 interaction promotes Rac1 signaling and metastatic
breast tumor cell invasion and migration. J Cell Biol. 150:177–191.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H, Thiele J and Vardiman JW: WHO Classification of
Tumours of Haematopoietic and Lymphoid Tissue (4th). Lyon, France:
IARC Press. 179–350. 2008.
|
|
20
|
Bierman PJ, Harris N and Armitage JO:
Non-Hodgkin's lymphoma. Cecil Medicine. Goldman L and Ausiello D:
(23rd). (Philadelphia). Saunders Elsevier. 1408–1425. 2008.
|
|
21
|
Komabayashi Y, Kishibe K, Nagato T, Ueda
S, Takahara M and Harabuchi Y: Downregulation of miR-15a due to
LMP1 promotes cell proliferation and predicts poor prognosis in
nasal NK/T-cell lymphoma. Am J Hematol. 89:25–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang HB, Zhan R, Wu SQ, Xu ZZ and Fan LP:
Expression of MCL-1 and microRNA-29a in extranodal NK/T-cell
lymphoma tissue. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 21:95–98.
2013.(In Chinese). PubMed/NCBI
|
|
23
|
Motsch N, Alles J, Imig J, Zhu J, Barth S,
Reineke T, Tinguely M, Cogliatti S, Dueck A, Meister G, et al:
MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell
lymphomas by deep sequencing. PLoS One. 7:e421932012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ti HJ, Nong L, Wang W, Zhang S and Li T:
Expression of microRNA in extranodal NK/T cell lymphoma, nasal
type. Zhonghua Bing Li Xue Za Zhi. 40:610–615. 2011.(In Chinese).
PubMed/NCBI
|
|
25
|
Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM,
Heo DS and Kim CW: MicroRNA-146a downregulates NFκB activity via
targeting TRAF6 and functions as a tumor suppressor having strong
prognostic implications in NK/T cell lymphoma. Clin Cancer Res.
17:4761–4771. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu S, Deng S, Ma Q, Zhang T, Jia C, Zhuo
D, Yang F, Wei J, Wang L, Dykxhoorn DM, et al: MicroRNA-10A* and
MicroRNA-21 modulate endothelial progenitor cell senescence via
suppressing high-mobility group A2. Circ Res. 112:152–164. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Long MJ, Wu FX, Li P, Liu M, Li X and Tang
H: MicroRNA-10a targets CHL1 and promotes cell growth, migration
and invasion in human cervical cancer cells. Cancer Lett.
324:186–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ohuchida K, Mizumoto K, Lin C, Yamaguchi
H, Ohtsuka T, Sato N, Toma H, Nakamura M, Nagai E, Hashizume M and
Tanaka M: MicroRNA-10a is overexpressed in human pancreatic cancer
and involved in its invasiveness partially via suppression of the
HOXA1 gene. Ann Surg Oncol. 19:2394–2402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X,
Guo X, He S and Chen R: Transcriptional inhibiton of Hoxd4
expression by miRNA-10a in human breast cancer cells. BMC Mol Biol.
10:122009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grady WM, Parkin RK, Mitchell PS, Lee JH,
Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz
AM, et al: Epigenetic silencing of the intronic microRNA
hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene.
27:3880–3888. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haapa-Paananen S, Chen P, Hellström K,
Kohonen P, Hautaniemi S, Kallioniemi O and Perälä M: Functional
profiling of precursor MicroRNAs identifies MicroRNAs essential for
glioma proliferation. PLoS One. 8:e609302013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Buffa FM, Camps C, Winchester L, Snell CE,
Gee HE, Sheldon H, Taylor M, Harris AL and Ragoussis J:
MicroRNA-associated progression pathways and potential therapeutic
targets identified by integrated mRNA and microRNA expression
profiling in breast cancer. Cancer Res. 71:5635–5645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu L, Liang YN, Luo XQ, Liu XD and Guo HX:
Association of miRNAs expression profiles with prognosis and
relapse in childhood acute lymphoblastic leukemia. Zhonghua Xue Ye
Xue Za Zhi. 32:178–181. 2011.(In Chinese). PubMed/NCBI
|
|
34
|
Ballabio E, Mitchell T, van Kester MS,
Taylor S, Dunlop HM, Chi J, Tosi I, Vermeer MH, Tramonti D,
Saunders NJ, et al: MicroRNA expression in Sezary syndrome:
Identification, function, and diagnostic potential. Blood.
116:1105–1113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu Y, Li XF, Yang JH, Liao XY and Chen YZ:
microRNAs expression profile in acute promyelocytic leukemia cell
differentiation induced by all-trans retinoic acid and arsenic
trioxide. Zhonghua Xue Ye Xue Za Zhi. 33:546–551. 2012.(In
Chinese). PubMed/NCBI
|
|
36
|
Leivonen SK, Sahlberg KK, Mäkelä R, Due
EU, Kallioniemi O, Børresen-Dale AL and Perälä M: High-throughput
screens identify microRNAs essential for HER2 positive breast
cancer cell growth. Mol Oncol. 8:93–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu
R, Pan Z, Kang T and Huang W: MicroRNA-342 inhibits colorectal
cancer cell proliferation and invasion by directly targeting DNA
methyltransferase 1. Carcinogenesis. 32:1033–1042. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
He YJ, Wu JZ, Ji MH, Ma T, Qiao EQ, Ma R
and Tang JH: miR-342 is associated with estrogen receptor-α
expression and response to tamoxifen in breast cancer. Exp Ther
Med. 5:813–818. 2013.PubMed/NCBI
|
|
39
|
Kim CH, Kim HK, Rettig RL, Kim J, Lee ET,
Aprelikova O, Choi IJ, Munroe DJ and Green JE: miRNA signature
associated with outcome of gastric cancer patients following
chemotherapy. BMC Med Genomics. 4:792011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu S, Huang S, Ding J, Zhao Y, Liang L,
Liu T, Zhan R and He X: Multiple microRNAs modulate p21Cip1/Waf1
expression by directly targeting its 3 untranslated region.
Oncogene. 29:2302–2308. 2010. View Article : Google Scholar : PubMed/NCBI
|