Introduction
Hepatocellular carcinoma (HCC) is the fifth most
frequent cancer worldwide and is also the third leading cause of
cancer-related mortality (1). The
World Health Organization has estimated that almost 56,400 new
cases of HCC occur globally per year (2), and the incidence is markedly higher in
men than in women. The highest liver cancer rates are observed in
developing countries, particularly in East Asia and Malaysia, South
Africa and Sub-Saharan Africa, whereas rates are lower in Europe,
North and South America, Australia and New Zealand (3). HCC may be induced by a number of risk
factors, including chronic infection with hepatitis B virus (HBV)
or hepatitis C virus (HCV), hepatic cirrhosis, alcoholic liver
disease and exposure to aflatoxins (4).
Despite recent advances in surgical treatments and
chemoradiotherapy, aggressive metastasis and early recurrence still
result in a high mortality rate among HCC patients (5). To date, a number of molecular markers
and signaling pathways have been identified to be associated with
HCC oncogenesis, progression, recurrence and survival, including
tumor protein P53 (TP53), K-ras mutation (6,7), HBx,
Notch 1, Glypican-3 and osteopontin overexpression (7–10),
Wnt/β-catenin and the phosphoinositide 3-kinase/protein kinase
B/mammalian target of rapamycin signaling pathways (11,12).
However, the exact mechanism of HCC has not been fully established.
Therefore, a better understanding of the molecular mechanisms
underlying HCC may contribute to the development of novel
strategies for prediction, diagnosis and therapy.
Long non-coding RNAs (lncRNAs) are
non-protein-coding transcripts of >200 nucleotides (13). A recent study found that the human
genome consists of at least four times more lncRNA sequences than
coding RNA sequences (14). The
majority of lncRNAs demonstrated to be functional are involved in
the regulation of gene transcription, post-transcriptional
regulation and epigenetic regulation, such as genomic imprinting
and X-inactivation (15–17), and are closely associated with
numerous human diseases, including cancer (18,19). The
lncRNA HOX transcript antisense RNA (HOTAIR), a human gene located
on chromosome 12, is expressed from the HOXC locus and is important
in the transcriptional regulation of various genes (20). The 5′ end of HOTAIR interacts with and
induces the genome-wide retargeting of Polycomb Repressive Complex
2 (PRC2), which combines with HOTAIR to silence the transcription
of the HOXC locus, and promotes metastasis of breast cancer by
silencing multiple metastasis suppressor genes (21). The 3′ end of HOTAIR interacts with
lysine-specific histone demethylase 1A (22). Furthermore, recent studies have
reported that HOTAIR is highly expressed and associated with poor
prognosis in a number of types of cancer, including breast cancer,
epithelial ovarian cancer, pancreatic cancer, colorectal cancer,
non-small-cell lung cancer and endometrial carcinoma (21,23–27).
The objective of the current study was to
investigate the expression of HOTAIR in HCC and to further explore
its clinical significance and molecular mechanisms. Paired fresh
HCC samples and adjacent normal tissues were used to examine the
expression level of HOTAIR and analyze the association between
HOTAIR expression and clinicopathological characteristics.
Furthermore, a lentivirus-mediated RNA interference method was
utilized to investigate the role and molecular mechanism of HOTAIR
in HCC progression.
Materials and methods
Patients and tissue samples
The patients enrolled in the study were diagnosed
with primary HCC and underwent partial liver resection between
January and September 2012 at Chinese People's Liberation Army
(PLA) General Hospital (Beijing, China). A total of 60 paired
samples of HCC and non-cancerous tissue were obtained from the
resected tumors and adjacent normal liver tissues of the patients,
and were immediately frozen in liquid nitrogen and stored at −80°C
until use. This study was approved by the ethics committee of
Chinese PLA General Hospital (LREC 2012/40) and the protocol was
conducted with the prior written informed consent of all patients.
All samples were confirmed independently by two pathologists, and
the clinicopathological characteristics were documented and are
described in detail in Table I.
 | Table I.Association between HOTAIR expression
and clinicopathological parameters in hepatocellular carcinoma
patients (n=60). |
Table I.
Association between HOTAIR expression
and clinicopathological parameters in hepatocellular carcinoma
patients (n=60).
|
|
| HOTAIR expression,
n |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | n | High (n=36) | Low (n=24) | χ2 | P-value |
|---|
| Gender |
|
|
|
0.082 | 0.775 |
|
Male | 42 | 25 | 17 |
|
Female | 18 | 10 | 8 |
| Age (years) |
|
|
|
0.866 | 0.352 |
|
<50 | 22 | 10 | 12 |
|
≥50 | 38 | 22 | 16 |
| Tumor size
(cm) |
|
|
|
0.431 | 0.512 |
| ≤5 | 22 | 12 | 10 |
|
|
|
>5 | 38 | 24 | 14 |
|
|
| Tumor number |
|
|
|
3.386 | 0.066 |
|
Solitary | 42 | 22 | 20 |
|
|
|
Multiple | 18 | 14 | 4 |
|
|
| Serum α-fetoprotein
(µg/l) |
|
|
|
2.188 | 0.139 |
|
<400 | 32 | 22 | 10 |
|
|
|
≥400 | 28 | 14 | 14 |
|
|
| Peritumoral
tissue |
|
|
|
3.103 | 0.078 |
|
Non-cirrhotic | 2 | 0 | 2 |
|
|
|
Cirrhotic | 58 | 36 | 22 |
|
|
| Tumor
differentiation |
|
|
| 12.198 | 0.002a |
| Well | 6 | 0 | 6 |
|
|
|
Moderate | 28 | 16 | 12 |
|
|
|
Poor | 26 | 20 | 6 |
|
|
| TNM stage |
|
|
|
6.389 | 0.094 |
| I | 12 | 4 | 8 |
|
|
| II | 6 | 4 | 2 |
|
|
|
III | 30 | 18 | 12 |
|
|
| IV | 12 | 10 | 2 |
|
|
| Vessel embolus |
|
|
|
3.403 | 0.065 |
|
Negative | 48 | 26 | 22 |
|
|
|
Positive | 12 | 10 | 2 |
|
|
| Metastasis |
|
|
| 10.000 | 0.002a |
|
Negative | 48 | 24 | 24 |
|
|
|
Positive | 12 | 12 | 0 |
|
|
| Early recurrence
(<2 years) |
|
|
| 11.250 | 0.001a |
|
Negative | 40 | 18 | 22 |
|
|
|
Positive | 20 | 18 | 2 |
|
|
RNA extraction, reverse transcription
(RT) and quantitative (q) polymerase chain reaction (PCR)
Total RNA from frozen HCC and paired non-cancerous
tissues or cell lines were extracted with the Ultrapure RNA Kit
(CWBio, Co., Ltd., Beijing, China) according to the manufacturer's
instructions. cDNA was synthesized by reverse transcribing the
total RNA using a HiFi-MMLV cDNA Kit (CWBio, Co., Ltd.). The
expression level of HOTAIR was detected by qPCR using the Ultra
SYBR Mixture with ROX (CWBio, Co., Ltd.) and ABI7500 system
(Applied Biosystems Life Technologies, Foster City, CA, USA).
Template cDNA (2 µl) was mixed with 25µl 2X UltraSYBR Mixture with
ROX, 2 µl primers and RNase-free water. β-actin was used as an
internal control and HOTAIR values were normalized to β-actin. The
primer sequences (Invitrogen; Thermo Fisher Scientific, Waltham,
MA, USA) used were as follows: HOTAIR forward,
5′-GCAGTAGAAAAATAGACATAGGAGA-3′, and reverse,
5′-ATAGCAGGAGGAAGTTCAGGCATTG-3′; β-actin forward,
5′-ACTTAGTTGCGTTACACCCTT-3′, and reverse, 5′-GTCACCTTCACCGTTCCA-3′.
HOTAIR cDNA was amplified under the following conditions: Initital
denaturation at 95°C for 5 min followed by 30 cycles of
denaturation at 95°C for 30 sec and primer annealing at 55°C for 30
sec, with a final extension step at 72°C for 60 sec. Expression
levels were calculated using the 2−ΔΔCT method (28) and were normalized to that of the
housekeeping β-actin gene.
HOTAIR siRNA lentiviral expression
vector
The RNA interference sequence for human HOTAIR (Gene
ID, 100124700) was obtained from a previous article (21). The small interfering RNA (siRNA)
sequences were as follows: HOTAIR, 5′-UAACAAGACCAGAGAGCUGUU-3′;
negative control (NC), 5′-TTCTCCGAACGTGTCACGT-3′. The
oligonucleotides encoding short hairpin RNA (shRNA) were then
constructed and synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China) and annealed into double strands by Annealing
Buffer for RNA Oligos (Beyotime Institute of Biotechnology, Haimen,
China). Following digestion with BamHI and EcoRI
restriction endonucleases (TransGen Biotech, Inc., Beijing, China),
the double stranded DNA molecules were inserted into pGCSi-neo-GFP
lentiviral vector (lentiviral plasmid and packaging vectors were
provided by Dr Xiao-Lei Li from the Department of Molecular Biology
of Chinese PLA General Hospital). All of the constructed plasmids
were confirmed by DNA sequencing.
pGCSi-neo-GFP-HOTAIR-shRNA/NC-shRNA plasmid DNAs, along with
packaging vectors, were transiently transfected into HEK293T
(American Type Culture Collection, Manassas, VA, USA) cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific) according
to the manufacturer's instructions. At 48 h after transfection,
supernatants containing lentivirus were collected and purified by
ultracentrifugation at 70,000 × g at 4°C for 2 h. The titer of the
lentivirus was detected using Lentivirus-Associated HIV p24 ELISA
Kit (Cell Biolabs, Inc., San Diego, CA, USA).
Cell culture and lentiviral
infection
Human liver cancer HepG2 cells were purchased from
the American Type Culture Collection and maintained in the research
center from the Department of Gastroenterology in Chinese PLA
General Hospital. Cells were maintained in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific) supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific)
and penicillin-streptomycin (MP Biomedicals, Santa Ana, CA, USA) at
37°C in an atmosphere of 5% CO2. For transfection,
well-cultured cells were seeded into a 6-well plate at a density of
1×105 cells/well for 24 h. Subsequently, lentivirus containing
shRNA targeting HOTAIR or NC shRNA was added into the medium and
incubated for 24 h. After replacing the culture medium of each
well, cells were incubated for a further 48 h. qPCR was performed
to detect the interference efficiency of the HOTAIR shRNA.
MTT assay
For cell proliferation assays, HepG2 cells stably
transfected with HOTAIR shRNA or NC shRNA were digested with 0.25%
trypsin (Boster Inc., Wuhan, China), seeded into 96-well plates at
a density of 1,000 cells/well in a final volume of 100 µl, and
maintained in DMEM supplemented with 10% FBS. At different time
points (24 h, 48 h, 72 h, 4 days, 5 days, 6 days or 7 days after
plating), 10 µl MTT solution (Sigma-Aldrich, St. Louis, MO, USA)
was added to each well, and cells were incubated for an additional
4 h at 37°C. The blue formazan crystals were dissolved in 100 µl
dimethyl sulfoxide (Origen Bioedical, Austin, TX, USA), and the
absorbance was measured at 490 nm using a microplate reader
(ELx800; BioTek Instuments, Inc., Winooski, VT, USA).
Transwell assay
A Transwell assay was performed to assess the
invasiveness of HepG2 cells by using chambers with an 8.0 µm
transparent polyethylene terephthalate membrane in 24-well plates
(Corning Incorporated, Corning, New York, NY, USA). Cells (2×105
per chamber) were seeded into the upper chambers in 200 µl
serum-free DMEM, and 500 µl of DMEM with 10% FBS was added to each
lower chamber. Three duplicate wells were performed for each group.
After 12 h, the unfiltered cells at the top surface of the membrane
were gently removed with cotton swabs. The cells that had passed
through the filters were fixed in methanol for 15 min and stained
with hematoxylin (OriGene China, Beijing, China) for 20 min,
air-dried and photographed (Olympus Stream Image Analysis Software;
Olympus Corporation, Tokyo, Japan). The invasive cells were counted
under a microscope (magnification, ×200; CX31; Olympus Corporation)
in five randomly selected visual fields.
Xenograft model
To examine the ability of tumor formation in
vivo, the cell lines stably transfected with HOTAIR shRNA or NC
shRNA were injected into nude mice. Ten four-week-old BALB/c male
nude mice were purchased from the Vital River Laboratories
(Beijing, China) and randomly assigned to two groups. All nude mice
were bred and maintained at the the Vital River Laboratories
(Beijing, China) under specific pathogen-free conditions. The
research program, objective and animal use protocol were reviewed
and approved by the Animal Ethical and Welfare Committee of
Tsinghua University. Cell suspension (100 µl) containing 1×107
HepG2-HOTAIR-shRNA or HepG2-NC-shRNA cells was injected
subcutaneously into the back of each nude mouse. The tumor size and
weight of the mice were measured every 4 days from the 8th day
following injection. On the 28th day following injection, all mice
in the two groups were sacrificed by cervical dislocation and the
primary tumors were removed from each mouse. Tumor size and tumor
weight were measured and recorded. The tumor volume was calculated
according to the following formula: Tumor volume = length ×
width2 × π/6 (29).
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was performed to assess the
mRNA expression levels of Wnt and β-catenin in tumor tissues from
nude mice. The methods for RNA extraction and RT were performed
using the aforementioned method. 2X Taq MasterMix (CWBio, Co.,
Ltd.) was used to amplify the cDNA. The PCR cycling parameters (30
cycles) were as follows: Predenaturation (95°C, 5 min),
denaturation (95°C, 30 sec), annealing (55°C, 30 sec) and extension
(72°C, 60 sec). GAPDH was used as an internal control. The RNA
primers (Invitrogen; Thermo Fisher Scientific) used in the PCR were
as follows: Wnt forward, 5′-GGAGTTGTATTTGCCATCACCAGGG-3′, and
reverse, 5′-ATGCGCGGGCAAATTTGATCCCATA-3′; β-catenin forward,
5′-ACAAGCCACAAGATTACAAGAAACGG-3′, and reverse,
5′-CCACCAGAGTGAAAAGAACGATAGCT-3′; GAPDH forward,
5′-TGGAGTCTACTGGCGTCTT-3′, and reverse,
5′-TGTCATATTTCTCGTGGTTCA-3′. The PCR products were assessed by 1.5%
agarose gel electrophoresis. Expression levels were calculated
using the 2−ΔΔCT method (28) and were normalized to that of the
housekeeping β-actin gene
Statistical analysis
All statistical analyses were performed using SPSS
version 20.0 statistical software package (IBM SPSS, Armonk, NY,
USA) or GraphPad Prism version 5.0 (GraphPad Software, Inc., La
Jolla, CA, USA). A χ2 test was used to assess the
association between HOTAIR expression level or tumor recurrence and
clinicopathological characteristics. Comparisons of quantitative
data between two groups were analyzed by an independent-samples
t-test. Spearman's correlation test was used to analyze the
correlation between HOTAIR expression level and clinicopathological
factors. All experiments were repeated independently three times
and data are presented as the mean ± standard deviation. P<0.05
was considered to indicate statistical significance.
Results
Increased expression of HOTAIR in
HCC
To examine the expression level of the lncRNA HOTAIR
in HCC and its association with clinical progression of HCC, HOTAIR
expression was detected in 60 paired fresh HCC tissues and adjacent
non-cancerous tissues using qPCR. The results revealed that HOTAIR
expression was increased at least 2-fold relative to that in
non-cancerous samples in 36 (60%) HCC samples; the mean expression
level of HOTAIR in HCC samples was 5.7-fold higher than the mean
level in the adjacent non-cancerous tissue samples (P=0.014,
t-test; Fig. 1A).
Correlation between HOTAIR expression
and clinicopathological parameters of HCC
The association between HOTAIR expression and the
clinicopathological characteristics of patients with HCC was
assessed (Table I). Significant
associations were identified between the HOTAIR expression and a
number of clinicopathological parameters that represent higher
tumor burdens, including poor tumor differentiation (P=0.002),
metastasis (P=0.002) and early recurrence (within 2 years;
P=0.001). Statistical analyses revealed no association between
HOTAIR expression and tumor size, tumor number, serum α-fetoprotein
level, cirrhosis, tumor-node-metastasis (TNM) stage (30) or the presence of vessel emboli.
Spearman's rank correlation coefficient analysis revealed that a
high expression level of HOTAIR was strongly correlated with tumor
differentiation (r=0.391, P=0.002), TNM stage (r=0.293, P=0.023),
metastasis (r=0.408, P=0.001) and early recurrence (r=0.433,
P=0.001) (Table II). Taken together,
these observations indicated that increased HOTAIR expression is
associated with HCC progression and early recurrence.
 | Table II.Spearman rank correlation coefficient
analysis between HOTAIR expression level and clinicopathological
factors. |
Table II.
Spearman rank correlation coefficient
analysis between HOTAIR expression level and clinicopathological
factors.
|
| Relative HOTAIR
expression |
|---|
|
|
|
|---|
| Characteristic | Correlation
coefficient | P-value |
|---|
| Tumor size
(cm) | 0.085 | 0.520 |
| Tumor number | 0.238 | 0.068 |
| Serum α-fetoprotein
(µg/l) | −0.191 | 0.144 |
| Peritumoral tissue
cirrhosis | 0.227 | 0.081 |
| Tumor
differentiation | 0.391 | 0.002a |
| TNM stage | 0.293 | 0.023a |
| Vessel embolus | 0.238 | 0.067 |
| Metastasis | 0.408 | 0.001a |
| Early recurrence
(<2 years) | 0.433 | 0.001a |
Downregulation of HOTAIR inhibits
proliferation of HepG2 cells
As the clinical data indicated that the expression
level of HOTAIR was closely associated with human HCC progression,
the effect of HOTAIR on the proliferation of liver cancer cells was
investigated. The constructed lentiviral vectors designated
siHOTAIR and siNC were transfected into HepG2 liver cancer cells,
and the interference efficiency was detected by qPCR following
transfection for 72 h. As shown in Fig.
1B, the expression level of HOTAIR was significantly
downregulated by 95% in siHOTAIR cells compared with NC cells
(P<0.001).
An MTT assay was performed to examine the effect of
HOTAIR on liver cancer cell proliferation. The results revealed
that the cell proliferative ability of siHOTAIR stably transfected
cells was markedly inhibited compared to the NC cells (Fig. 1C). These data indicate that HOTAIR is
important in liver cancer cell proliferation in vitro.
Inhibition of HOTAIR reduces
invasiveness of HepG2 cells
A Transwell assay was used to assess the effect of
HOTAIR on the invasive ability of liver cancer cells. The results
revealed that the number of cells that passed through the filters
was significantly decreased in the siHOTAIR group compared with
that of siNC group (Fig. 1D).
Statistical analysis indicated that the difference between two
groups was significant (P=0.049). This finding demonstrated that
increased expression of HOTAIR may enhance liver cancer cell
invasiveness in vitro.
HOTAIR expression regulates the
tumorigenicity of HepG2 cells
To further determine whether HOTAIR expression could
promote HCC progression and enhance the tumorigenicity of liver
cancer cells, an in vivo tumor model was used. Using a shRNA
lentiviral knockdown system, two stable cell lines, HepG2-siHOTAIR
and HepG2-siNC, were generated and injected subcutaneously into the
backs of nude mice (n=5 per group). Tumor volumes were measured
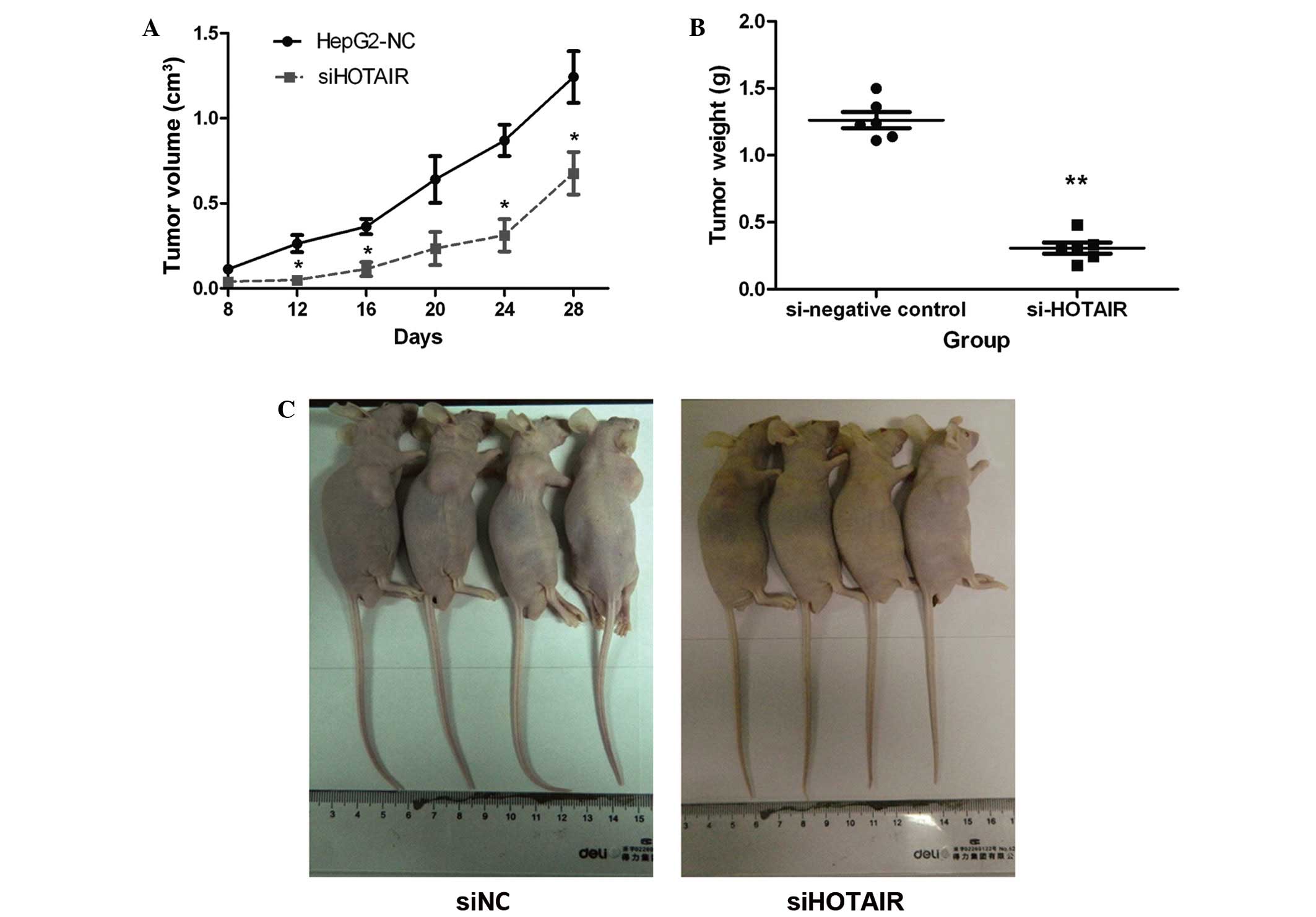
every 4 days until the 28th day following injection. As shown in
Fig. 2A, tumors from HepG2-siNC cells
grew faster than tumors formed from siHOTAIR cells. In addition,
the mean tumor weight in HepG2-siHOTAIR group was significantly
lower compared with that from the HepG2-siNC group (Fig. 2B). Four mice from each group were
photographed and tumor size in the siHOTAIR group was observed to
be markedly smaller than that in the NC group (Fig. 2C). These in vivo results were
consistent with the findings of the in vitro experiments,
and suggested that HOTAIR regulates liver cancer cell
proliferation, invasion and the tumorigenic ability, and may play
an important role in HCC progression.
HOTAIR regulates Wnt/β-catenin mRNA
expression
Tumor models of the nude mice from the two groups (5
per group) were used to detect the mRNA expression levels of Wnt
and β-catenin under the condition of HOTAIR inhibition using
semi-quantitative RT-PCR. The interference efficiency of HOTAIR in
these tumor models was confirmed by qPCR (Fig. 3A). As shown in Fig. 3B, in the siHOTAIR group, the
expression levels of Wnt and β-catenin were markedly reduced
compared with those of the NC group. These data indicate that
HOTAIR may regulate Wnt/β-catenin expression alone or in
combination with other molecules by an unknown mechanism that
merits further study.
Discussion
HCC is a significant health problem worldwide, with
high rates of incidence and mortality (2). Accumulative, long-term interactions
between environmental and genetic factors lead to the occurrence
and progression of HCC (31).
Although a number of tumor suppressor genes and oncogenes,
including KRAS, PTEN and TP53, have been
identified to be involved in HCC progression (6,7,32), the molecular mechanism of HCC is not
completely understood. In recent years, the focus of tumor research
has expanded to investigate the potential tumor suppressive or
oncogenic functions of lncRNAs (33,34).
HOTAIR is an oncogenic lncRNA that has been found to
interact with PRC2 to epigenetically regulate chromatin state and
multiple target genes (21).
Recently, accumulating evidence has demonstrated that HOTAIR is
dysregulated in various types of cancer. Gupta et al
(21) reported that HOTAIR is highly
expressed in metastatic breast cancer and its high expression in
primary breast tumors is a significant predictor of subsequent
metastasis and mortality (21). In a
similar manner, increased HOTAIR expression may also indicate poor
prognosis and promote metastasis in non-small cell lung cancer
(25), epithelial ovarian cancer
(27) and colon cancer (23).
In the current study, the expression level of HOTAIR
in HCC tissues versus adjacent non-cancerous tissues was examined
by quantitative RT-PCR, and its clinical implications were
investigated. The results revealed that HOTAIR expression was at
least 2-fold higher in 60% of HCC samples, and the extent of the
increase ranged from 2- to 46-fold (mean, 5.7-fold). With regard to
clinicopathological significance, HOTAIR expression levels were
closely associated with tumor differentiation (P=0.002), metastasis
(P=0.002) and early recurrence (P=0.001). Furthermore, a Spearman's
rank correlation coefficient analysis revealed that high HOTAIR
expression was linked to poor tumor differentiation (r=0.391,
P=0.002) and advanced TNM stage (r=0.293, P=0.023) and predicted a
greater tendency for metastasis (r=0.408, P=0.001) and early
recurrence (r=0.433, P=0.001). From these data, we hypothesize that
HOTAIR participates in the development and progression of HCC. As
certain clinicopathological characteristics, including tumor
differentiation, TNM stage, metastasis and recurrence, always
indicate poor prognosis, HOTAIR may be a predictor of poor
prognosis in HCC patients, warranting further research.
Previous reports have suggested that HOTAIR may
regulate cell proliferation, migration and invasion in a variety of
tumor cell types (35–37). In endometrial carcinoma, the
overexpression of HOTAIR increases the malignant potential of tumor
cells in vitro and in vivo (24). The knockdown of HOTAIR lncRNA
suppresses tumor invasion in gastric cancer cells and reverses the
epithelial-mesenchymal transition (38). To explore the biological function of
HOTAIR in HCC progression, a lentivirus-mediated shRNA expression
system capable of interfering with HOTAIR expression in HepG2 cells
was developed in the present study. The data demonstrated that
knockdown of HOTAIR significantly suppressed the proliferation and
invasion of HepG2 cells in vitro, and effectively reduced
the oncogenicity of liver cancer cells HepG2 in vivo. Taken
together, these data indicate that high levels of HOTAIR may
promote the progression of liver cancer cells; this mechanism
remains to be explored.
The Wnt/β-catenin signaling pathway is a group of
signal transduction pathways, which was first identified for its
role in carcinogenesis and has been demonstrated to promote tumor
development in multiple types of human cancer. Ge et al
(39) found that HOTAIR is able to
directly reduce the expression of Wnt inhibitory factor 1,
activating the Wnt/β-catenin signaling pathway; this clarified one
of the molecular mechanisms underlying progression and metastasis
in esophageal squamous cell carcinoma, and may represent a novel
therapeutic target in these patients (39). The present study examined the
expression of Wnt and β-catenin in neoplasms of the nude mice from
two groups. The results revealed that the mRNA expression level of
Wnt and β-catenin were repressed when HOTAIR was silenced. Emerging
evidence has demonstrated that the Wnt/β-catenin signaling pathway
plays a key role during HCC genesis and development, and may be a
therapeutic target in human HCC (40,41). We
hypothesize that HOTAIR may influence carcinogenesis partially
through activating and cooperating with the Wnt/β-catenin signaling
molecules in HCC. However, further research is necessary to clarify
the exact mechanism of HOTAIR in HCC.
In conclusion, the present study demonstrated that
HOTAIR expression is upregulated in the majority of HCC tissues,
and is closely associated with tumor differentiation, metastasis
and early recurrence in HCC patients. Furthermore, the
overexpression of HOTAIR promotes HCC progression partly by
activating the Wnt/β-catenin signaling pathway. Thus,
downregulating HOTAIR by interference may serve as a promising
therapeutic strategy for the treatment of HCC.
Acknowledgements
The authors would like to thank their colleagues
from the Department of Molecular Biology and the Department of
Gastroenterology of Chinese PLA General Hospital for their
technical support.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosch FX, Ribes J, Diaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127(5 Suppl 1): S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Turdean S, Gurzu S, Turcu M, Voidazan S
and Sin A: Current data in clinicopathological characteristics of
primary hepatic tumors. Rom J Morphol Embryol. 53(Suppl 3):
719–724. 2012.PubMed/NCBI
|
|
4
|
Severi T, van Malenstein H, Verslype C and
van Pelt JF: Tumor initiation and progression in hepatocellular
carcinoma: Risk factors, classification and therapeutic targets.
Acta pharmacol Sin. 31:1409–1420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang ZY: Hepatocellular carcinoma
surgery-review of the past and prospects for the 21st century. J
Surg Oncol. 91:95–96. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qi LN, Bai T, Chen ZS, Wu FX, Chen YY, De
Xiang B, Peng T, Han ZG and Li LQ: The p53 mutation spectrum in
hepatocellular carcinoma from Guangxi, China: Role of chronic
hepatitis B virus infection and aflatoxin B1 exposure. Liver Int.
35:999–1009. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ye H, Zhang C, Wang BJ, Tan XH, Zhang WP,
Teng Y and Yang X: Synergistic function of Kras mutation and HBx in
initiation and progression of hepatocellular carcinoma in mice.
Oncogene. 33:5133–5138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Gao JZ, Du JL and Wei LX: Prognostic
and clinicopathological significance of glypican-3 overexpression
in hepatocellular carcinoma: A meta-analysis. World J
Gastroenterol. 20:6336–6344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Q, Wang R, Wang Y, et al: Notch1 is a
potential therapeutic target for the treatment of human hepatitis B
virus X protein-associated hepatocellular carcinoma. Oncol Rep.
31:933–939. 2014.PubMed/NCBI
|
|
10
|
Zhang CH, Xu GL, Jia WD, Ge YS, Li JS, Ma
JL and Ren WH: Prognostic significance of osteopontin in
hepatocellular carcinoma: A meta-analysis. Int J Cancer.
130:2685–2692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie L, Jiang H and Wu F: Role of
Wnt/β-catenin signaling pathway in promoting tumorigenesis of
hepatocellular carcinoma. Nan Fang Yi Ke Da Xue Xue Bao.
34:913–917. 2014.(In Chinese). PubMed/NCBI
|
|
12
|
Janku F, Kaseb AO, Tsimberidou AM, Wolff
RA and Kurzrock R: Identification of novel therapeutic targets in
the PI3K/AKT/mTOR pathway in hepatocellular carcinoma using
targeted next generation sequencing. Oncotarget. 5:3012–3022. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Perkel JM: Visiting ‘noncodarnia’.
Biotechniques. 54:301, 303–304. 2013.
|
|
14
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koerner MV, Pauler FM, Huang R and Barlow
DP: The function of non-coding RNAs in genomic imprinting.
Development. 136:1771–1783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Costanzi C and Pehrson JR: Histone
macroH2A1 is concentrated in the inactive X chromosome of female
mammals. Nature. 393:599–601. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell
Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reis EM, Nakaya HI, Louro R, Canavez FC,
Flatschart AV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa
F, et al: Antisense intronic non-coding RNA levels correlate to the
degree of tumor differentiation in prostate cancer. Oncogene.
23:6684–6692. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu ZH, Wang XL, Tang HM, Jiang T, Chen J,
Lu S, Qiu GQ, Peng ZH and Yan DW: Long non-coding RNA HOTAIR is a
powerful predictor of metastasis and poor prognosis and is
associated with epithelial-mesenchymal transition in colon cancer.
Oncol Rep. 32:395–402. 2014.PubMed/NCBI
|
|
24
|
Huang J, Ke P, Guo L, Wang W, Tan H, Liang
Y and Yao S: Lentivirus-mediated RNA interference targeting the
long noncoding RNA HOTAIR inhibits proliferation and invasion of
endometrial carcinoma cells in vitro and in vivo. Int J Gynecol
Cancer. 24:635–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW,
Jin HY, Zhang Y, Li Q and Hua KQ: Overexpression of long non-coding
RNA HOTAIR predicts poor patient prognosis and promotes tumor
metastasis in epithelial ovarian cancer. Gynecol Oncol.
134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shao R, Bao S, Bai X, Blanchette C,
Anderson RM, Dang T, Gishizky ML, Marks JR and Wang XF: Acquired
expression of periostin by human breast cancers promotes tumor
angiogenesis through up-regulation of vascular endothelial growth
factor receptor 2 expression. Mol Cell Biol. 24:3992–4003. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu TH, Wang CC, Huang CC, Chen CL, Hung
CH, Chen CH, Wang JH, Lu SN, Lee CM, Changchien CS and Tai MH:
Down-regulation of tumor suppressor gene PTEN, overexpression of
p53, plus high proliferating cell nuclear antigen index predict
poor patient outcome of hepatocellular carcinoma after resection.
Oncol Rep. 18:1417–1426. 2007.PubMed/NCBI
|
|
33
|
Niland CN, Merry CR and Khalil AM:
Emerging roles for long non-coding RNAs in cancer and neurological
disorders. Front Genet. 3:252012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Wu Z, Mei Q, Li X, Guo M, Fu X and
Han W: Long non-coding RNA HOTAIR, a driver of malignancy, predicts
negative prognosis and exhibits oncogenic activity in oesophageal
squamous cell carcinoma. Br J Cancer. 109:2266–2278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ,
Huang L, Yu PF and Cheng XD: Knockdown of long non-coding RNA
HOTAIR suppresses tumor invasion and reverses
epithelial-mesenchymal transition in gastric cancer. Int J Biol
Sci. 9:587–597. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY,
Xue WQ, Chen YB, Zhang Y and Jia WH: HOTAIR, a prognostic factor in
esophageal squamous cell carcinoma, inhibits WIF-1 expression and
activates Wnt pathway. Cancer Sci. 104:1675–1682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qu B, Liu BR, Du YJ, Chen J, Cheng YQ, Xu
W and Wang XH: Wnt/β-catenin signaling pathway may regulate the
expression of angiogenic growth factors in hepatocellular
carcinoma. Oncol Lett. 7:1175–1178. 2014.PubMed/NCBI
|
|
41
|
Dahmani R, Just PA and Perret C: The
Wnt/β-catenin pathway as a therapeutic target in human
hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
35:709–713. 2011. View Article : Google Scholar : PubMed/NCBI
|