Introduction
Previously, studies categorized non-coding (nc)RNAs
as short ncRNA, mid-size ncRNA, and long ncRNA (lncRNA) by their
lengths (1,2). In addition, lncRNAs are subdivided by
function, loci and post-transcriptional modification. lncRNAs have
been operationally defined as transcripts that are produced by RNA
polymerase II (Pol II), which are broadly classified as transcripts
>200 nucleotides in length (3,4).
Transcriptome analyses have revealed that 70–90% of the mammalian
genome was transcribed, but only 1–2% may encode proteins (5). Additionally, lncRNAs that are localized
to the nucleus possess stronger secondary structures. Therefore,
knockdown of lncRNAs may not be sufficiently effective at evoking a
phenotype and uncovering the physiological function of the lncRNA
(5–8).
The regulation of mRNA decay in the cytoplasm is crucial for
controlling the abundance of cellular transcripts and the levels of
protein expression. Therefore, the regulation of lncRNA decay in
the nucleus is considered to be important for biological function
(9). Overall, lncRNAs are important
in the programming and regulation of the mammalian genome. The
expression of lncRNAs is associated with numerous human diseases,
including cancer (10).
The prevention of 20% of cancers in the United
States alone would result in 300,000 fewer new cases annually
(11). The growing knowledge of
cancer treatment reveals methods to intercept cancers by novel,
active approaches. Cancer results in high mortality and morbidity
globally, largely due to the complex, heterogeneous nature of the
disease and the lack of biomarkers for early diagnosis. A
proteomics study of cancer identified functional proteins and drove
the transformation of malignancy, identified biomarkers to detect
early-stage cancer, determined therapy efficacy, identified novel
drug targets and ultimately developed personalized medicine
(12). Since tumor formation is a
multistep process, normal cells evolve progressively to the
neoplastic stage. Therefore, normal cells may acquire particular
capacities that enable them to become tumorigenic. Over the last
decade, remarkable progress was made in the field of cancer
research, which led to a better understanding of cancer therapy
(13).
Notably, metastasis associated lung adenocarcinoma
transcript-1 (MALAT-1) is reported in numerous studies. For
example, MALAT-1 was detected in the cerebellum of human alcoholics
(14). However, MALAT-1 is a typical
multifunctional gene that is important in a wide array of cancers,
including bladder cancer (15,16),
gallbladder carcinoma (GBC) (17,18),
hepatocellular carcinoma (HCC) (19)
and lung (20,21) and gastric cancer (GC) (22). Therefore, understanding the regulation
function of MALAT-1 within the context of cancer is of considerable
significance. The present study focuses on the recent advances in
the role of MALAT-1 in diverse cancers, as MALAT-1 possesses a
typical multifunctional lncRNA (Table
I).
 | Table I.The expression of MALAT-1 in cancer.
MALAT-1 was expressed in all samples. |
Table I.
The expression of MALAT-1 in cancer.
MALAT-1 was expressed in all samples.
| Sample | Function in
tumorigenesis | References |
|---|
| HeLa, CaSki, SiHa
and HCC94 cells | Oncogene | (32,49) |
| Gallbladder
carcinoma tissues | Oncogene | (22) |
| Bladder cancer
tissues | Oncogene | (16,25) |
| Mesenchymal stem
cells | Oncogene | (75) |
| SGC-7901, MKN-45
and SUN-16 cells | Biomarker | (5) |
| Human colorectal
cancer tissues and LoVo and SW620 cells | Biomarker | (20) |
| Liver cancer
tissues and hepatocellular carcinoma cells | Biomarker | (6,11) |
| Neuroblastoma
SK-N-SH cells | Oncogene | (75) |
| Non-small cell lung
cancer cells | Biomarker | (20,36) |
| Pancreatic ductal
adenocarcinoma tissues | Biomarker | (38) |
| Melanoma
tissues | Oncogene | (76) |
| Bladder urothelial
carcinoma cells | Biomarker | (3) |
| Brain metastasis
lung cancer cells | Biomarker | (10) |
| Colorectal cancer
tissues | Biomarker | (48) |
| Gastric cancer
tissues | Biomarker | (14) |
Overview of MALAT-1
MALAT-1 is generally defined as a lncRNA that
consists of >8,000 nt and is coded by chromosome 11q13. MALAT-1
transcripts are upregulated in a number of human carcinomas, highly
expressed in numerous cancer types, including bladder cancer,
gallbladder carcinoma, liver cancer, melanoma, colorectal cancer
and gastric cancer, and associated with metastasis. MALAT-1 lacks a
significant open reading frame; therefore, it may not translate
proteins in vitro (23,24).
Studies have indicated that MALAT-1 possesses a distinct sequence
or secondary structure that directs localization to nuclear
speckles in human tumor cells (25,26).
Additionally, MALAT-1 expression may be abrogated using zinc finger
nucleases (5). Notably, the
quantitative loss of MALAT-1 did not affect proliferation, cell
cycle progression or nuclear architecture in human lung or liver
cancer cells (8). The MALAT-1 mouse
model did not reveal any evident phenotype or histological
abnormalities compared with wild-type animals. In addition, the
loss of abundant nuclear MALAT-1 is compatible with cell viability
and normal development (8). Previous
studies have demonstrated that MALAT-1 interacts with pre-mRNA
splicing factors, including the serine- and arginine-rich (SR)
family of proteins (27–29). In detail, MALAT-1 may regulate
numerous biological processes, including cancer cell migration,
synapse formation, cell cycle progression and response to serum
stimulation. However, MALAT-1 function becomes apparent only in
specific cell types, such as metastatic cancer cells, and under
particular conditions (27). MALAT-1
controls cell cycle progression by modulating the oncogenic
transcription factor Myb-related protein B (B-MYB). B-MYB is a
transcription factor that is required for the transcription of a
large number of genes involved in mitotic progression (30).
Xu et al divided MALAT-1 into five fragments.
The fragment located in the 3′ end (6,918–8,441 nt), was pivotal in
the biological processes of cell proliferation, migration and
invasion in colorectal cancer (CRC) SW620 and SW480 cells (31). Notably, MALAT-1 exhibited
substantially high expression levels in HeLa and MCF-7 cells and
was clearly downregulated in a dose-dependent manner. According to
the decline, MALAT-1 demonstrates a statistically significant
dose-dependent decrease in human melanoma (BLM)-treated HeLa cells,
but not in BLM-treated MCF-7 cells or irradiated cells (32). The 3′ end of MALAT-1 is transcribed by
Pol II and formed by the cleavage of ribonuclease P (RNase P). The
3′ ends form a novel triple-helical structure that is essential for
stimulating translation, the stability of the RNA and supporting
export to the cytoplasm (33).
Notably, another study indicated that the 3′ end processing
mechanism of MALAT-1 may yield a stable nuclear-retained ncRNA with
a short poly (A) tail-like moiety and a small tRNA-like cytoplasmic
RNA (34). MALAT-1 has been
implicated in the regulation of mRNA splicing and expression
(35). In addition, MALAT-1 is
associated with prostate cancer progression, including
castration-resistant prostate cancer (CRPC) (36). Tee et al observed that MALAT-1
induced neuroblastoma cell migration and invasion (37). MALAT-1 may also specifically regulate
gene expression, but not alternative splicing in lung cancer cells
(38). Additionally, MALAT-1 is
abundantly expressed in the SK-N-SH cell line under normal culture
conditions and the activation of the oxytocin receptor resulted in
a significant increase of MALAT-1 expression (39). The results of an additional study of
the 5′ end of the MALAT-1 transcript indicated that an alternate
transcription initiation site was used in the neuroblastoma cell
line, which resulted in a shorter transcript compared with the
previously reported transcript in lung cancer cells (39).
Previous studies have demonstrated that MALAT-1 is a
highly conserved transcript that regulates the expression of
metastasis-associated genes (38,40).
Analysis of the nuclear/cytoplasmic distribution of MALAT-1 during
the cell cycle reveals a distinct profile, which demonstrates a
profound enrichment in the G2/M phase in HeLa cells (28). The heterogeneous nuclear
ribonucleoprotein (hnRNP) C protein is of particular interest, due
to the RNA-binding capability of the protein and the reported
cytoplasmic translocation in the G2/M phase (41). Compared with the insufficient binding
capacity at the 5′ end of the transcript, strong binding to the
hnRNP C protein has been indicated in other regions of MALAT-1
(42). The function of MALAT-1 in the
cell cycle may be regulated by facilitating the cytoplasmic
translocation of the hnRNP C protein. However, in a previous study,
MALAT-1 silencing only compromised the cytoplasmic translocation
and not the expression of the hnRNP C protein (42).
Molecular targets of MALAT-1
At present, numerous studies have reported the
antitumor role of MALAT-1 in cancer development (Table II). In addition, high expression of
MALAT-1 has been indicated in cervical (30) and lung cancer (43). MALAT-1 is involved in the pathogenesis
of cancers through the regulation of carcinoma-associated signaling
pathways, including the mitogen-activated protein kinase (MAPK)
signaling pathway (17). Tripathi
et al showed that the specific deletion of MALAT-1 in human
osteosarcoma cells led to the activation of p53 (30). MALAT-1-depleted cells demonstrated
cell cycle defects that were sensitive to the p53 levels, which
indicates that p53 is a main downstream mediator for MALAT-1
activity. In MALAT-1-depleted cells, replication of the S-phase was
decreased and the cell population of G1 and G2/M cells was
increased. In addition, MALAT-1 was involved in cervical cancer
cell growth, cell cycle progression and invasion. The accumulation
of evidence indicated that the downregulation of MALAT-1 may induce
the expression of caspase-3, caspase-8 and B-cell lymphoma 2
(Bcl-2)-associated × protein. In addition, MALAT-1 may suppress the
expression of Bcl-2 and B-cell lymphoma-extra large. Overall, these
results suggested that MALAT-1 may act as a therapeutic target in
the prevention of human cervical cancer (44).
 | Table II.Targets of metastasis associated lung
adenocarcinoma transcript 1 in cancer. |
Table II.
Targets of metastasis associated lung
adenocarcinoma transcript 1 in cancer.
| Cellular
system | Modulators | Target
molecules | Biological
consequences | References |
|---|
| HeLa cells |
| p53 | Cell cycle
arrest | (30) |
| HeLa cells | SRSF1 |
| Alternative
splicing | (28) |
| Human cervical
cancer | Caspase-3,
caspase-8, Bax, Bcl-2 and Bcl-xL |
| Cell growth, cell
cycle progression and invasion | (44) |
| Gallbladder
carcinoma |
| ERK/MAPK | Proliferation and
metastasis | (17) |
| Gallbladder
carcinoma |
| Cyclin D1, cyclin E
and CDK6 | Cell migration and
proliferation | (49) |
| Bladder cancer | TGF-β |
|
Epithelial-mesenchymal transition and
tumor metastasis | (15) |
| Bladder cancer |
| Wnt/β-catenin |
Epithelial-mesenchymal transition and
sequent cell migration | (16) |
| Human
osteosarcoma | Myc-6 |
| Cell growth | (48) |
| Myeloma |
| Sp1, LTBP3 and
TGF-β | Therapeutic target
biomarker | (49) |
| Gastric cancer |
| SF2/ASF | Cell
proliferation | (22) |
| Colorectal
cancer |
| SFPQ | Tumor metastasis,
prognosis and therapeutic target | (54) |
| Esophageal squamous
cell carcinoma | SOX17 |
| Cell growth and
migration | (55) |
| Liver cancer | YAP, SRSF1 |
| Transwell
mobility | (19) |
| Neuroblastoma | CREB |
| Tumor marker | (39) |
| Human and mouse
lung cancer | ASOs |
|
| (38) |
The knockdown of MALAT-1 in GBC cell lines may
significantly inhibit the proliferation and metastasis of the GBC
cells in vitro and in vivo. In addition, the
extracellular signal-regulated kinase/MAPK signaling pathway may be
inactivated in the GBC cell lines following MALAT-1 knockdown,
which indicates that MALAT-1 may act as an oncogenic lncRNA that
promotes the proliferation and metastasis of GBC (17). Other studies indicate that
transforming growth factor-β (TGF-β) induces MALAT-1 expression and
epithelial-mesenchymal transition (EMT) in bladder cancer cells
(15,16,45,46). In
addition, the inhibition of MALAT-1 or suppressor of zeste 12
suppressed the migratory and invasive properties induced by TGF-β
(15). A previous study revealed that
the Wnt signaling pathway demonstrated particularly close links
with EMT (47). Immunostaining
analysis showed that MALAT-1-siRNA treatment significantly
decreased the nuclear accumulation of β-catenin. Therefore, MALAT-1
promotes EMT and sequent cell migration by activating the
Wnt/β-catenin signaling pathway in bladder cancer cells (16).
MALAT-1 is recognized as an oncogene, as it
regulates the alternative splicing of endogenous target genes that
are involved in cancer (40).
Taniguchi et al indicated that Myc-6 acts as a nuclear
sequence-specific transcription factor, which specifically binds to
the theoretical target site that is located within the 5′-upstream
region of MALAT-1 (48). In addition,
Myc-6 exposure led to a dose-dependent decrease in the expression
level of MALAT-1 in human osteosarcoma MG63 cells. Multiple
serine/arginine-rich splicing factor 1 (SRSF1) proteins bound
specifically and directly to the 5′ end of MALAT-1. Additionally,
MALAT-1 regulated alternative splicing by modulating the levels of
active SR proteins (28). Li et
al considered the potential significance that MALAT-1 promotes
the activation effect of the key transcription factor Sp1 on the
latent TGF-β-binding protein 3 (LTBP3) promoter by modulating the
recruitment of Sp1 to the LTBP3 gene, which regulates the
bioavailability of TGF-β, particularly in mesenchymal stem cells
from myeloma patients (49). MALAT-1
may promote cell migration and decrease cell proliferation in CaSki
cells. Notably, MALAT-1 may increase cell proliferation by
upregulating cyclin D1, cyclin E and cluster of differentiation K6
(18). The study conducted by Wang
et al indicated that MALAT-1 was overexpressed in GC cells
(22). Additional studies revealed
that MALAT-1 may induce a particularly high expression of SF2/ASF,
an important member of the SR protein family, in the nucleolus. The
depression of SF2/ASF induced poor cell proliferation in a similar
way to MALAT-1 depletion; however, no significant effect on the
cell proliferation depression or the loss of nuclear distribution
of SF2/ASF was observed with low MALAT-1 expression. Therefore,
MALAT-1 promotes cell proliferation in GC cells by recruiting
SF2/ASF.
In tumorigenesis studies, MALAT-1 is one of several
important splicing factor proline/glutamine-rich (SFPQ)-binding
RNAs (50–53). SFPQs may also be termed PTB-associated
splicing factors. Additionally, MALAT-1 may competitively bind to
SFPQ to release SFPQ from the SFPQ/polypyrimidine tract binding
protein 2 (PTBP2) complex in CRC LoVo cells. SFPQs, not PTBP2
proteins, are critical in the regulatory effect (54). In esophageal squamous cell carcinoma,
MALAT-1 is suppressed by sex determining region Y (SRY)-box 17 via
SRY binding-mediated transcriptional regulation (55).
Cancer metastasis has been indicated as the major
cause of cancer recurrence and tumor-associated mortality. In a
previous study, antisense oligonucleotides (ASOs) were designed to
potently target MALAT-1. The altered ASOs effectively reduced
MALAT-1 expression compared with the control ASO (38). A study conducted by Wang et al
indicated an interaction between the Yes-associated protein (YAP)
and SRSF1 by Angiomotin regulated MALAT-1 (19). The MALAT-1-mediated tumorigenesis in
HCC provided a novel mechanism for YAP regulation of gene
expression at the transcriptional and post-transcriptional stages.
The cyclic AMP-responsive element binding protein was indicated to
bind to the proximal promoter of MALAT-1 in the human neuroblastoma
SK-N-SH cell line (39). These
results demonstrated that MALAT-1 expression may be modulated by
the extracellular stimulation of tumor cells (Fig. 1).
 | Figure 1.Target function of MALAT-1 in cancer.
MALAT-1 acts as an oncogene by targeting significant tumor
suppressor genes, including SF2/ASF, SFPQ, SRSF1, cyclin D1, cyclin
E and CDK6. Overexpression of MALAT-1 increases cell proliferation
through the activation of ERK/MAPK, p53, Wnt/β-catenin, caspase-3,
caspase-8, the Bax signaling pathway and the cell cycle. MALAT-1
may suppress the expression of Bcl-2 and Bcl-xL. The expression of
MALAT-1 is suppressed by Myc-6 and SOX17. TGF-β induces MALAT-1
expression. CREB binds to the proximal promoter of the MALAT-1.
MALAT-1, metastasis associated lung adenocarcinoma transcript 1;
SF2/ASF, splicing factor 2/alternative splicing factor; SFPQ,
splicing factor proline/glutamine rich; SRSF1, serine/arginine-rich
splicing factor 1; CDK6, cyclin-dependent kinase 6; ERK/MAPK,
extracellular signal-regulated kinase/mitogen-activated protein
kinase pathway; Bax, Bcl-2-like protein 4; Bcl-2, B-cell lymphoma
2; Bcl-xL, B-cell lymphoma-extra large; TGF-β, transforming growth
factor; CREB, cAMP response element-binding protein; Sox17, sex
determining region Y-box 17; p53, tumor protein p53; Sp1/LTBP3,
transcription factor Sp1/latent transforming growth factor β
binding protein 3. |
The identification of MALAT-1 target genes that are
involved in tumor development may aid the development of novel
therapeutic strategies for tumor intervention.
MALAT-1 and epigenetic regulation
As aforementioned, the upregulation of MALAT-1 has
been described in several types of human tumors. MALAT-1 silencing
or gene therapy may be effective therapeutic approaches for human
tumors (10,38,56). At
present, epigenetic mechanisms are important in the regulation of
gene expression. The role of MALAT-1 is being elucidated in the
epigenetic field. For example, post-translational histone
modifications and RNA-based mechanisms, including those controlled
by microRNAs, are significant mechanisms of the epigenetic
regulation of gene expression. Overall, MALAT-1 may affect cancer
development (57,58). Notably, Jumonji C-domain-containing
protein (JMJD)1A belongs to the JMJD family. Tee et al
observed that the anti-JMJD1A antibody efficiently
immunoprecipitated the MALAT-1 gene core promoter (37). In addtion, JMJD1A increased MALAT1
gene transcription by demethylating histone H3K9. Addtional studies
revealed that JMJD1A increased MALAT-1 expression by directly
binding to the MALAT-1 gene promoter, which lead to histone H3K9
demethylation by activating JMJD1A gene transcription (59,60).
Overall, the findings suggest that the development of more potent
JMJD1A/MALAT-1 inhibitors may be used for the prevention of tumor
metastasis (37). The
methylation/demethylation cycle of Polycomb 2 (Pc2), a component of
the polycomb repressive complex 1, is responsible for the physical
relocation of growth control genes from Polycomb bodies (PcGs).
Methylated Pc2 may demonstrate an antimitogenic signal, but
unmethylated Pc2 is essential for physiological growth control,
gene expression and cell proliferation. Additionally, a sequence
analysis of 94 various clones identified that taurine upregulated 1
(TUG1) repressed cell cycle genes, and was the most enriched RNA
that interacted with methylated Pc2 (61). TUG1 localizes to PcGs and interacts
with the methylated form of Pc2. However, MALAT-1 resides in
nuclear speckles and only interacts with the unmethylated Pc2
protein (62). MALAT-1 silencing has
been indicated to inhibit cell growth in HeLa cells, even in the
presence of growth signals. The promoters that are associated with
MALAT-1 in these domains exhibit high levels of activating the
methylation of Lys36 at histone H3 trimethylation (H3K36me3) and
H3K4me3 marks. This finding may be facilitated by the interaction
of MALAT-1 with lysine demethylase 1 and SET nuclear proto-oncogene
(SET) domain-containing 2. Histone methyltransferases and
demethylases are associated with transcriptional activation and
pre-mRNA splicing factors (63,64). In
summary, the results of previous studies may influence the
chromatin modifications and accessibility of target gene loci.
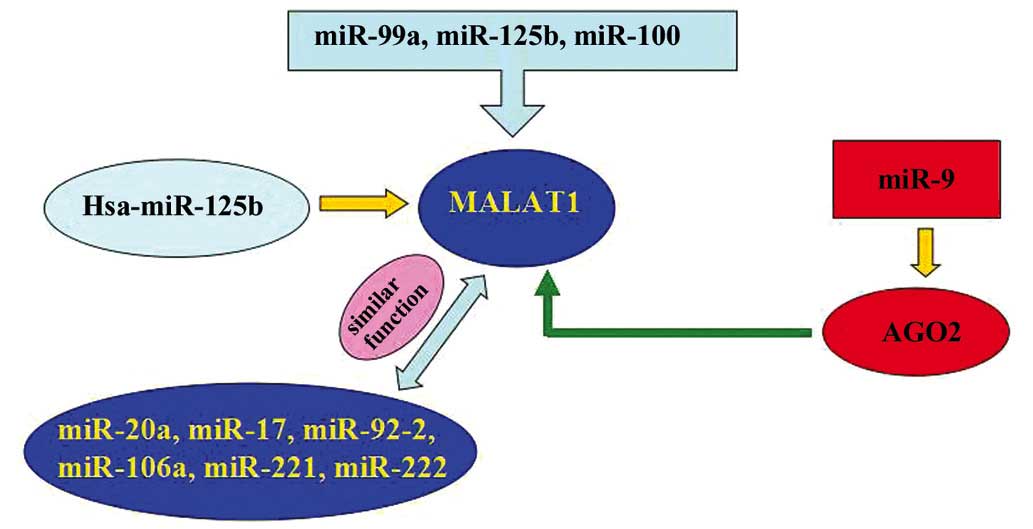
Evidently, microRNA-125b (miR-125b), miR-99a and
miR-100 are overexpressed in vincristine-resistant acute
lymphoblastic leukemia (ALL). Co-expression of these miRNAs
resulted in the downregulation of DNA nucleotidylexotransferase,
nuclear casein kinase and cyclin-dependent kinase substrate 1,
MALAT-1, small nuclear ribonucleoprotein polypeptide E, PNO1
protein, SET, kinesin-1 heavy chain, phosphoribosyl pyrophosphate
synthetase 2, and ribosomal proteins S11, L38 and L23a. One study
indicated that 7 of these genes, including MALAT-1, demonstrated
decreased expression in the vincristine-resistant ALL cells of
children (65). Another study showed
that certain miRNAs, including miR-20a, −17, −92-2, −106a, −221 and
−222, and chr19 ncRNA, which is similar to MALAT-1, may contribute
to c-myc induced mouse mammary tumors (66). miR-9 may regulate the expression of
MALAT-1. In addition, miR-9 may target AGO2-mediated regulation of
MALAT-1 in the nucleus (67). Han
et al hypothesized that Hsa-miR-125b is downregulated in
bladder cancer compared with matched normal urothelium (68). However, sirtuin (SIRT)7 and MALAT-1
were upregulated in bladder cancer compared with matched normal
urothelium. SIRT7 is identified as a crucial regulator of
mitochondrial homeostasis. Additionally, high-grade and high-stage
carcinomas demonstrated increased expression levels of SIRT7 and
MALAT-1. However, additional studies indicated lower Hsa-miR-125b
expression levels of SIRT7 and MALAT-1 compared with low-grade and
low-stage carcinomas. In addition, Hsa-miR-125b mimics demonstrated
markedly inhibited MALAT-1 expression levels in T24 and 5637 cells,
whereas hsa-miR-125b inhibitors demonstrated markedly increased
MALAT-1 expression levels. Notably, hsa-miR-125b binding sites
within MALAT-1 were functional (Fig.
2).
Potential clinical applications of
MALAT-1
Studies on MALAT-1 are, at present, at an early
stage, and no extensive clinical studies on the expression of
MALAT-1 in cancer tissues have been reported. In addition, the
functional roles and associations of MALAT-1 with cancer are
unclear. Therefore, MALAT-1 may be expected to potentially function
as novel biomarkers in the diagnosis, prognosis, metastasis and the
prediction of responses to therapy in solid tumors.
Numerous studies have assessed the biology of
MALAT-1 in cancer (15–22). The role of MALAT-1 in cellular
transformation suggests that MALAT-1 has the potential to function
as a biomarker and a target for novel therapeutic approaches in
multiple myeloma (MM). Previous studies have suggested that MALAT-1
expression in B-cell malignancies is decreased compared with solid
tumors (27,69). Another study demonstrated
significantly lower MALAT-1 expression levels in the plasma of MM
patients (70).
The study conducted by Jiang et al identified
the correlation between MALAT-1 and human papillomavirus (HPV)
(18). In this study, 64 cases of
clinical cervical squamous cell carcinoma (SCC) samples were
collected. MALAT-1 was identified in 6/18 cases in HPV-positive
cervical normal cells and 14/22 cases in HPV-positive cervical
lesion specimens. Therefore, these results suggested that HPV was
an important factor that led to that activation of MALAT-1 in
cervical SCC.
MALAT-1 was previously indicated to demonstrate
extremely high expression levels in the established human non-small
cell lung cancer (NSCLC) A549 and HTB58 cell lines. In addition,
the downregulation of MALAT-1 decreased tumor growth in
vivo. Human MALAT-1 transcripts increased the migration
potential in the mouse fibroblast NIH3T3 cell line, and the
downregulation of MALAT-1 decreased the migration potential and
tumor growth in human NSCLC A549 cells in vivo. Overall, for
SCC, increased expression of MALAT-1 may be associated with a poor
prognosis (20). Notably, the A549
MALAT-1 wild-type cells and the two knockout (KO) cell lines with
the lowest MALAT-1 expression, KO2 and KO3, were injected into the
tail vein of nude mice, and the formation of the lung tumor nodules
was analyzed 2 months later. The results of this pharmacological
study suggested that MALAT-1 is required for effective tumor nodule
formation in vivo, and therefore affects the metastatic
process in lung cancer (38).
In order to identify tumor-associated MALAT-1 and to
determine the correlation of the transcript with pancreatic duct
adenocarcinoma (PDAC), Weber et al identified that MALAT-1
complies with the key characteristics of diagnostic biomarkers,
including minimal invasiveness, high specificity and robustness
(21). Alternatively, MALAT-1 may be
applied as a complementary biomarker within a panel, in order to
improve the entire diagnostic performance. The results of the study
also showed that MALAT-1 expression levels are upregulated in
pancreatic cancer tissues compared with adjacent noncancerous
controls. Consistently, a higher expression level of MALAT-1 was
identified in all seven pancreatic cancer cell lines relative to
the human pancreatic ductal epithelial cell. Function analysis
revealed that the downregulation of MALAT-1 may inhibit tumor cell
proliferation and decrease cell migration and invasion. The
underlying mechanisms are possibly involved in inducing G2/M cell
cycle arrest, promoting cell apoptosis, suppressing EMT and
reducing cancer stem-like properties. Therefore, the accumulation
of evidence indicates that MALAT-1 may act as an oncogenic lncRNA
that is involved in the malignant phenotype of pancreatic cancer.
Importantly, MALAT-1 may be used as a potential therapeutic target
(71). Another study indicated that
the overexpression of MALAT-1, the tumor location and nerve
invasion were independent predictors of disease-specific survival
of PDAC (56).
MALAT-1 overexpression has been reported to predict
the recurrence of HCC following liver transplantation (LT). MALAT-1
rs619586 was associated with a decreased HCC risk, with a
borderline significance (P=0.057). Larger studies are required in
order to clarify the associations between rs619586 in MALAT-1 and
HCC risk (72). Notably, a
consistently higher expression level of MALAT-1 was identified in
all nine liver cancer cell lines, relative to the normal liver LO2
cell line. The association study indicated that the overexpression
of MALAT-1 did not exhibit significant correlations with the
pathological features of age, gender, tumor size, histological
differentiation or portal vein tumor thrombi (73). More importantly, the multivariate
regression analysis revealed that the overexpression of MALAT-1 may
be a novel independent predictor of recurrence-free survival in HCC
patients following LT. In addition, MALAT-1 silencing in HCC may be
a potential anticancer therapy to prevent tumor recurrence
following orthotopic liver transplantations (74). However, MALAT-1 is overexpressed in
hepatoblastomas (HPBL) compared with HCC, as HCC and HPBL have
distinct patterns of gene expression (75). Subsequent studies have provided
evidence about the importance of MALAT-1 in liver cell
proliferation, which was confirmed by the finding of arrested liver
cell proliferation in response to partial MALAT-siRNA mediated
knockdown (40,76,77).
The expression level of MALAT-1 was significantly
increased in melanoma tissues compared with paired adjacent normal
tissues. Although no statistical difference of MALAT-1 expression
was observed between melanomas with or without lymph node
metastasis, the expression level of MALAT-1 was affected by the
metastatic status of the tumor in melanoma. In addition, the
migration of A-375 cells transfected with MALAT-1-siRNA was
estimated using a Transwell assay. The cells with impaired
expression of MALAT-1 migrated less effectively through the
Transwell membrane. Therefore, the enhanced expression of MALAT-1
had the potential to affect cancer metastasis in melanomas. In
general, these findings indicated that the expression level of
MALAT-1 had the potential to be a prognostic indicator for the
metastasis of melanoma (78).
17β-Estradiol (E2) treatment to breast cell lines
causes MALAT-1 RNA to decrease in an estrogen receptor α
independent manner. This effect of E2 treatment is caused by
MALAT-1 transcriptional regulation of E2. Zhao et al
hypothesized that the effects of E2 treatment on breast cells are
achieved by regulating MALAT-1 (79).
However, MALAT-1 was downregulated in the cell culture, with the
cells exhibiting high metastatic potential for ovarian cancer
metastasis. The function of MALAT-1 may vary with various cancer
types and context (80).
Several reports indicate that MALAT-1 contributes to
the complex molecular mechanisms involved in the control of cell
growth, differentiation and motility. Therefore, MALAT-1 may be
important in the process of cancer metastasis. Han et al
observed that MALAT-1 is upregulated in bladder urothelial
carcinoma compared with the matched normal urothelium (81). A high expression level of MALAT-1 was
associated with high-grade and high-stage bladder urothelial
carcinoma. MALAT-1 silencing inhibited bladder urothelial carcinoma
cell growth, induced apoptosis and decreased cell motility in T24
and 5637 cells. In addition, MALAT-1 knockdown also inhibited tumor
metastasis (15). MALAT-1 inhibition
may represent a promising therapeutic option for the suppression of
bladder cancer progression. MALAT-1 expression levels were
significantly upregulated in the majority of bladder cancer tissues
compared with normal tissues. These data indicated that the
upregulation of MALAT-1 may be associated with the onset of bladder
cancer metastasis. In addition, MALAT-1 silencing impaired bladder
cancer cell migration (16).
A study conducted by Okugawa et al
illustrated that the expression levels of both MALAT-1 and HOX
transcript antisense RNA were significantly increased in GC tissues
compared with matching adjacent normal mucosa (82). Elevated MALAT-1 expression
significantly correlated with peritoneal dissemination.
Additionally, a difference in the levels of MALAT-1 was not evident
in the plasma in a comparison between the healthy controls and GC
patients (83).
The expression level of MALAT-1 was significantly
increased in brain metastasis samples compared with non-brain
metastasis samples. MALAT-1 was increased in a highly invasive
subline of brain metastasis lung cancer cells. Functional studies
indicated that MALAT-1 silencing inhibits a highly invasive subline
of brain metastasis lung cancer cell migration and metastasis by
inducing EMT (10).
A recent study indicated that the expression of
MALAT-1 was higher in human CRC tissues than adjacent normal
tissues (54). MALAT-1 upregulation
has been demonstrated in CRC tumors. In addition, the expression
levels of MALAT-1 in cancerous tissues were increased compared with
noncancerous tissues. Although no associations were identified
between MALAT-1 expression and the stages, including stage II and
III, in CRC patients, the expression of MALAT-1 was significantly
increased in male patients compared with female patients. In
particular, patients with a high level of MALAT-1 expression showed
significantly shorter disease-free survival (DFS) and overall
survival times (OS) compared with patients with low MALAT-1
expression. In addition, patients with perineural invasion
demonstrated significantly shorter DFS and OS times compared with
those without perineural invasion. Multivariate Cox regression
analysis indicated that MALAT-1 expression and perineural invasion
were predictors of poor prognosis regarding DFS in CRC patients
(84).
Another study investigated that MALAT-1 was
upregulated across all 8 CRPC samples, which has been implicated in
regulating mRNA splicing. The expression of MALAT-1 was also
recently indicated to be associated with prostate cancer
progression (36).
MALAT-1 and chemoresistance
MALAT-1 has been indicated to promote cell
proliferation, migration and inhibit cell apoptosis in numerous
cancer cells. The expression of MALAT-1 was reported to
dramatically decrease with the increasing concentration of
cisplatin and paclitaxel, which lengthened the treatment duration.
Cisplatin and paclitaxel target significant lncRNAs in laryngeal
squamous cell carcinoma (LSCC), which provides a novel molecular
target to treat LSCC patients and sets direction for the
development of novel drugs (85).
Lung cancer is the top cause of cancer-associated mortality
(86). One reason for this is the
development of resistance to the chemotherapy treatment. In
particular, cancer stem cells (CSCs) may escape treatment and
regenerate the bulk of the tumor. Gene expression analysis showed
that MALAT-1 was involved in tumor development and metastasis. In
addition, MALAT-1 was similarly induced in CSCs and
cisplatin-resistant H460 cells (87).
Conclusion and future perspectives
The identification of ncRNAs as important regulators
of gene expression improved the understanding of numerous
biological processes, including cancer cell migration, synapse
formation, cell cycle progression and response to serum
stimulation. The findings indicate that MALAT-1 is an important
novel modulator in the development of cancers. MALAT-1 may promote
cell proliferation, inhibit apoptosis, and enhance tumor growth in
numerous cancer cell lines, and therefore, may serve as an
excellent candidate for therapeutic intervention. High expression
levels of MALAT-1 are widely hypothesized to associate with the
prognosis and survival in clinical cancer patients. Notably, single
MALAT-1 detection has strong potential as an independent prognostic
factor in cancer patients. However, the applicability and
epigenetic regulation of MALAT-1 targeted strategies for the
clinical treatment of human tumors requires additional studies.
Additional studies are also required to elucidate
the mechanism of MALAT-1, the emerging targets in oncology and the
possible future directions for clinical applications. Therefore,
in-depth studies on the functions of MALAT-1 may lead to novel
diagnostic and therapeutic approaches.
References
|
1
|
Core LJ, Waterfall JJ and Lis JT: Nascent
RNA sequencing reveals widespread pausing and divergent initiation
at human promoters. Science. 322:1845–1848. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1 - a paradigm for long noncoding RNA function in cancer. J
Mol Med Berl. 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiu MT, Hu JW, Yin R and Xu L: Long
noncoding, RNA. An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gutschner T, Baas M and Diederichs S:
Noncoding RNA gene silencing through genomic integration of RNA
destabilizing elements using zinc finger nucleases. Genome Res.
21:1944–1954. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang B, Arun G, Mao YS, Lazar Z, Hung G,
Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C and Spector DL:
The lncRNA Malat1 is dispensable for mouse development but its
transcription plays a cis-regulatory role in the adult. Cell Rep.
2:111–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eißmann M, Gutschner T, Hämmerle M,
Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun
T, Zörnig M and Diederichs S: Loss of the abundant nuclear
non-coding RNA MALAT1 is compatible with life and development. RNA
Biol. 9:1076–1087. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tani H, Nakamura Y, Ijiri K and Akimitsu
N: Stability of MALAT-1, a nuclear long non-coding RNA in mammalian
cells, varies in various cancer cells. Drug Discov Ther. 4:235–239.
2010.PubMed/NCBI
|
|
10
|
Shen L, Chen L, Wang Y, Jiang X, Xia H and
Zhuang Z: Long noncoding RNA MALAT1 promotes brain metastasis by
inducing epithelial-mesenchymal transition in lung cancer. J
Neurooncol. 121:101–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Perera FP and Vineis P: Cancer. IARC Sci
Publ. 337–362. 2011.PubMed/NCBI
|
|
12
|
Tan HT, Lee YH and Chung MC: Cancer
proteomics. Mass Spectrom Rev. 31:583–605. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dillman RO: Cancer immunotherapy. Cancer
Biother Radiopharm. 26:1–64. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kryger R, Fan L, Wilce PA and Jaquet V:
MALAT-1, a non protein-coding RNA is upregulated in the cerebellum,
hippocampus and brain stem of human alcoholics. Alcohol.
46:629–634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding
Q, Weng H, Shu YJ, Liu TY, Jiang L, et al: MALAT1 promotes the
proliferation and metastasis of gallbladder cancer cells by
activating the ERK/MAPK pathway. Cancer Biol Ther. 15:806–814.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie
P, Zhou G and Li G: The role of MALAT1 correlates with HPV in
cervical cancer. Oncol Lett. 7:2135–2141. 2014.PubMed/NCBI
|
|
19
|
Wang J, Wang H, Zhang Y, Zhen N, Zhang L,
Qiao Y, Weng W, Liu X, Ma L, Xiao W, et al: Mutual inhibition
between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced
tumourigenesis in liver cancer. Cell Signal. 26:1048–1059. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weber DG, Johnen G, Casjens S, Bryk O,
Pesch B, Jöckel KH, Kollmeier J and Brüning T: Evaluation of long
noncoding RNA MALAT1 as a candidate blood-based biomarker for the
diagnosis of non-small cell lung cancer. BMC Res Notes. 6:5182013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Su L, Chen X, Li P, Cai Q, Yu B,
Liu B, Wu W and Zhu Z: MALAT1 promotes cell proliferation in
gastric cancer by recruiting SF2/ASF. Biomed Pharmacother.
68:557–564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rajaram V, Knezevich S, Bove KE, Perry A
and Pfeifer JD: DNA sequence of the translocation breakpoints in
undifferentiated embryonal sarcoma arising in mesenchymal hamartoma
of the liver harboring the t(11;19)(q11;q13.4) translocation. Genes
Chromosomes Cancer. 46:508–513. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyagawa R, Tano K, Mizuno R, Nakamura Y,
Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T and
Akimitsu N: Identification of cis- and trans-acting factors
involved in the localization of MALAT-1 noncoding RNA to nuclear
speckles. RNA. 18:738–751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hutchinson JN, Ensminger AW, Clemson CM,
Lynch CR, Lawrence JB and Chess A: A screen for nuclear transcripts
identifies two linked noncoding RNAs associated with SC35 splicing
domains. BMC Genomics. 8:392007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakagawa S, Ip JY, Shioi G, Tripathi V,
Zong X, Hirose T and Prasanth KV: Malat1 is not an essential
component of nuclear speckles in mice. RNA. 18:1487–1499. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Spector DL and Lamond AI: Nuclear
speckles. Cold Spring Harb Perspect Biol. 3:a0006462011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A and
Prasanth KV: Long noncoding RNA MALAT1 controls cell cycle
progression by regulating the expression of oncogenic transcription
factor B-MYB. PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: A long non-coding RNA and its important 3′ end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI
|
|
32
|
Özgür E, Mert U, Isin M, Okutan M, Dalay N
and Gezer U: Differential expression of long non-coding RNAs during
genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin
Exp Med. 13:119–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marzluff WF: Novel 3′ ends that support
translation. Genes Dev. 26:2457–2460. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wilusz JE, Freier SM and Spector DL: 3′
end processing of a long nuclear-retained noncoding RNA yields a
tRNA-like cytoplasmic RNA. Cell. 135:919–932. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tollervey JR, Curk T, Rogelj B, Briese M,
Cereda M, Kayikci M, Hortobágyi T, Nishimura AL, Župunski V, et al:
Characterising the RNA targets and position-dependent splicing
regulation by TDP-43. Nat Neurosci. 14:452–458. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sowalsky AG, Xia Z, Wang L, Zhao H, Chen
S, Bubley GJ, Balk SP and Li W: Whole transcriptome sequencing
reveals extensive unspliced mRNA in metastatic castration-resistant
prostate cancer. Mol Cancer Res. 13:98–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tee AE, Ling D, Nelson C, Atmadibrata B,
Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, et al: The
histone demethylase JMJD1A induces cell migration and invasion by
up-regulating the expression of the long noncoding RNA MALAT1.
Oncotarget. 5:1793–1804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koshimizu TA, Fujiwara Y, Sakai N, Shibata
K and Tsuchiya H: Oxytocin stimulates expression of a noncoding RNA
tumor marker in a human neuroblastoma cell line. Life Sci.
86:455–460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mohamadkhani A: Long noncoding RNAs in
interaction with RNA binding proteins in hepatocellular carcinoma.
Hepat Mon. 14:e187942014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schepens B, Tinton SA, Bruynooghe Y,
Parthoens E, Haegman M, Beyaert R and Cornelis S: A role for hnRNP
C1/C2 and Unr in internal initiation of translation during mitosis.
EMBO J. 26:158–169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang F, Yi F, Han X, Du Q and Liang Z:
MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett.
587:3175–3181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guffanti A, Iacono M, Pelucchi P, Kim N,
Soldà G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ,
et al: A transcriptional sketch of a primary human breast cancer by
454 deep sequencing. BMC Genomics. 10:1632009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guo F, Li Y, Liu Y, Wang J, Li Y and Li G:
Inhibition of metastasis-associated lung adenocarcinoma transcript
1 in CaSki human cervical cancer cells suppresses cell
proliferation and invasion. Acta Biochim Biophys Sin (Shanghai).
42:224–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wei H, Kamat AM, Aldousari S, Ye Y, Huang
M, Dinney CP and Wu X: Genetic variations in the transforming
growth factor beta pathway as predictors of bladder cancer risk.
PLoS One. 7:e517582012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Al-Azayzih A, Gao F, Goc A and Somanath
PR: TGFβ1 induces apoptosis in invasive prostate cancer and bladder
cancer cells via Akt-independent, p38 MAPK and JNK/SAPK-mediated
activation of caspases. Biochem Biophys Res Commun. 427:165–170.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu W, Ji J, Xu Y, Liu Y, Shi L, Liu Y, Lu
X, Zhao Y, Luo F, Wang B, et al: MicroRNA-191, by promoting the EMT
and increasing CSC-like properties, is involved in neoplastic and
metastatic properties of transformed human bronchial epithelial
cells. Mol Carcinog. 54(Suppl 1): E148–E161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Taniguchi M, Fujiwara K, Nakai Y, Ozaki T,
Koshikawa N, Toshio K, Kataba M, Oguni A, Matsuda H, Yoshida Y, et
al: Inhibition of malignant phenotypes of human osteosarcoma cells
by a gene silencer, a pyrrole-imidazole polyamide, which targets an
E-box motif. FEBS Open Bio. 4:328–334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li B, Chen P, Qu J, Shi L and Zhuang W, Fu
J, Li J, Zhang X, Sun Y and Zhuang W: Activation of LTBP3 gene by a
long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem
cells from multiple myeloma. J Biol Chem. 289:29365–29375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li L, Feng TT, Lian YY, Zhang GF, Garen A
and Song X: Role of human noncoding RNAs in the control of
tumorigenesis. Proc Natl Acad Sci USA. 106:12956–12961. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang G, Cui Y, Zhang G, Garen A and Song
X: Regulation of proto-oncogene transcription, cell proliferation,
and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA.
Proc Natl Acad Sci USA. 106:16794–16798. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sephton CF, Cenik C, Kucukural A, Dammer
EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, et al:
Identification of neuronal RNA targets of TDP-43-containing
ribonucleoprotein complexes. J Biol Chem. 286:1204–1215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rajesh C, Baker DK, Pierce AJ and Pittman
DL: The splicing-factor related protein SFPQ/PSF interacts with
RAD51D and is necessary for homology-directed repair and sister
chromatid cohesion. Nucleic Acids Res. 39:132–145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kuo IY, Wu CC, Chang JM, Huang YL, Lin CH,
Yan JJ, Sheu BS, Lu PJ, Chang WL, Lai WW and Wang YC: Low SOX17
expression is a prognostic factor and drives transcriptional
dysregulation and esophageal cancer progression. Int J Cancer.
135:563–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu JH, Chen G, Dang YW, Li CJ and Luo DZ:
Expression and prognostic significance of lncRNA MALAT1 in
pancreatic cancer tissues. Asian Pac J Cancer Prev. 15:2971–2977.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Abi Khalil C: The emerging role of
epigenetics in cardiovascular disease. Ther Adv Chronic Dis.
5:178–187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Asrih M and Steffens S: Emerging role of
epigenetics and miRNA in diabetic cardiomyopathy. Cardiovasc
Pathol. 22:117–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Park SJ, Kim JG, Son TG, Yi JM, Kim ND,
Yang K and Heo K: The histone demethylase JMJD1A regulates
adrenomedullin-mediated cell proliferation in hepatocellular
carcinoma under hypoxia. Biochem Biophys Res Commun. 434:722–727.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Osawa T, Tsuchida R, Muramatsu M,
Shimamura T, Wang F, Suehiro J, Kanki Y, Wada Y, Yuasa Y, Aburatani
H, et al: Inhibition of histone demethylase JMJD1A improves
anti-angiogenic therapy and reduces tumor-associated macrophages.
Cancer Res. 73:3019–3028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang L, Lin C, Liu W, Zhang J, Ohgi KA,
Grinstein JD, Dorrestein PC and Rosenfeld MG: NcRNA- and Pc2
methylation-dependent gene relocation between nuclear structures
mediates gene activation programs. Cell. 147:773–788. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Smith MA, Gesell T, Stadler PF and Mattick
JS: Widespread purifying selection on RNA structure in mammals.
Nucleic Acids Res. 41:8220–8236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt
KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG and
Fu XD: Enhancing nuclear receptor-induced transcription requires
nuclear motor and LSD1-dependent gene networking in interchromatin
granules. Proc Natl Acad Sci USA. 105:19199–19204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wagner EJ and Carpenter PB: Understanding
the language of Lys36 methylation at histone H3. Nat Rev Mol Cell
Biol. 13:115–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Moqadam Akbari F, Lange-Turenhout EA,
Ariës IM, Pieters R and den Boer ML: MiR-125b, miR-100 and miR-99a
co-regulate vincristine resistance in childhood acute lymphoblastic
leukemia. Leuk Res. 37:1315–1321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sun Y, Wu J, Wu SH, Thakur A, Bollig A,
Huang Y and Liao DJ: Expression profile of microRNAs in c-Myc
induced mouse mammary tumors. Breast Cancer Res Treat. 118:185–196.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Leucci E, Patella F, Waage J, Holmstrøm K,
Lindow M, Porse B, Kauppinen S and Lund AH: MicroRNA-9 targets the
long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep.
3:25352013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bernard D, Prasanth KV, Tripathi V,
Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L,
Coulpier F, et al: A long nuclear-retained non-coding RNA regulates
synaptogenesis by modulating gene expression. EMBO J. 29:3082–3093.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Isin M, Ozgur E, Cetin G, Erten N, Aktan
M, Gezer U and Dalay N: Investigation of circulating lncRNAs in
B-cell neoplasms. Clin Chim Acta. 431:255–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J,
Zhang Y, Chen J, Shen H and Hu Z: A genetic variant in long
non-coding RNA HULC contributes to risk of HBV-related
hepatocellular carcinoma in a Chinese population. PLoS One.
7:e351452012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li G, Zhang H, Wan X, Yang X, Zhu C, Wang
A, He L, Miao R, Chen S and Zhao H: Long noncoding RNA plays a key
role in metastasis and prognosis of hepatocellular carcinoma.
Biomed Res Int. 2014:7805212014.PubMed/NCBI
|
|
74
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Luo JH, Ren B, Keryanov S, Tseng GC, Rao
UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, et al:
Transcriptomic and genomic analysis of human hepatocellular
carcinomas and hepatoblastomas. Hepatology. 44:1012–1024. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Watts R, Ghozlan M, Hughey CC, Johnsen VL,
Shearer J and Hittel DS: Myostatin inhibits proliferation and
insulin-stimulated glucose uptake in mouse liver cells. Biochem
Cell Biol. 92:226–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
MALAT-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhao Z, Chen C, Liu Y and Wu C:
17β-Estradiol treatment inhibits breast cell proliferation,
migration and invasion by decreasing MALAT-1 RNA level. Biochem
Biophys Res Commun. 445:388–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu SP, Yang JX, Cao DY and Shen K:
Identification of differentially expressed long non-coding RNAs in
human ovarian cancer cells with different metastatic potentials.
Cancer Biol Med. 10:138–141. 2013.PubMed/NCBI
|
|
81
|
Han Y, Liu Y, Nie L, Gui Y and Cai Z:
Inducing cell proliferation inhibition, apoptosis, and motility
reduction by silencing long noncoding ribonucleic acid
metastasis-associated lung adenocarcinoma transcript 1 in
urothelial carcinoma of the bladder. Urology. 81:209e1–209.e7.
2013. View Article : Google Scholar
|
|
82
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR and
Goel A: Metastasis-associated long non-coding RNA drives gastric
cancer development and promotes peritoneal metastasis.
Carcinogenesis. 35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Arita T, Ichikawa D, Konishi H, Komatsu S,
Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T,
et al: Circulating long non-coding RNAs in plasma of patients with
gastric cancer. Anticancer Res. 33:3185–3193. 2013.PubMed/NCBI
|
|
84
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
85
|
Chen H, Xin Y, Zhou L, Huang JM, Tao L,
Cheng L and Tian J: Cisplatin and paclitaxel target significant
long noncoding RNAs in laryngeal squamous cell carcinoma. Med
Oncol. 31:2462014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hong SH, Park SJ, Lee S, Cho CS and Cho
MH: Aerosol gene delivery using viral vectors and cationic carriers
for in vivo lung cancer therapy. Expert Opin Drug Deliv.
12:977–991. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lopez-Ayllon BD, Moncho-Amor V, Abarrategi
A, de Ibañez Cáceres I, Castro-Carpeño J, Belda-Iniesta C, Perona R
and Sastre L: Cancer stem cells and cisplatin-resistant cells
isolated from non-small-lung cancer cell lines constitute related
cell populations. Cancer Med. 3:1099–1111. 2014. View Article : Google Scholar : PubMed/NCBI
|