Introduction
Human saliva is a complex biological fluid that
contains electrolytes, enzymes such as amylase and carbonic
anhydrase, proteins (including glycoproteins such as mucins and
proline-rich glycoproteins) and peptides such as statherin,
cystatins, histatins and proline-rich proteins (1).
Human saliva participates in important biological
functions in the mouth, and is essential for mastication and
digestion (2). Furthermore, it
protects the oral health by means of lysozymes, cystatins,
immunoglobulins and histatins present on the saliva, which prevent
the growth of microorganisms in the oral cavity (3).
The protein content of whole saliva derives from the
three major paired salivary glands, which comprise the
contralateral major (parotid, submandibular and sublingual) and
minor salivary glands (4). The
protein composition of whole saliva depends on the circadian
rhythm, diet, age, gender and physiological status of the
individual (5).
The study of the salivary proteome may aid to
identify all the proteins present in the saliva, and may detect
alterations in protein levels that occur in specific physiological
and possibly pathological conditions (6).
The classical proteomic approach for the study of
proteins is based on two-dimensional polyacrylamide gel
electrophoresis (2D-PAGE) to compare and identify differences in
the protein expression patterns between diseased and normal samples
(7,8).
Following 2D-PAGE fractionation and staining, the proteins of
interest are removed, digested (proteolytically or chemically) and
identified by matrix-assisted laser
desorption/ionization-time-of-flight/mass spectrometry
(MALDI-TOF/MS) (7,8). However, this approach presents several
limitations, since it is time consuming, expensive and requires
difficult and laborious tasks (7,8).
Proteomic analysis has recently benefited from the
introduction of surface-enhanced laser desorption/ionization
(SELDI)-TOF. SELDI-TOF/MS requires the use of chips, and
constitutes a simple and high-throughput technique to rapidly
identify a large number of differentially expressed peptides and
proteins in saliva samples, particularly proteins of low molecular
weight (<10 kDa) that are difficult to detect effectively with
other methods (9). In addition, since
SELDI-TOF/MS requires minimal quantities of a biological sample to
generate an accurate protein profile in a relatively short period
of time, it is suitable for mapping protein profiles in samples
derived from healthy and diseased individuals in order to identify
differential protein expression patterns between the two groups
(10). The results of a SELDI-TOF/MS
analysis are usually presented as a list of proteins that are
upregulated or downregulated in healthy versus diseased subjects
(11).
SELDI technology is also useful for the protein
profiling of a variety of complex biological tissues and fluids,
including serum, blood, plasma, intestinal fluid, urine, cell
lysates and cellular secretion products (12). The working principle and preparation
of the protein chip in SELDI are the same for every biological
fluid analyzed, whereas the pretreatment differs according to each
biological matrix (12).
SELDI-TOF/MS has revealed numerous novel biomarkers
for different types of cancer, including ovarian (13), breast (14), pancreatic (15) and prostate cancer (16). In addition, this technology has been
used for obtaining proteomic patterns for the diagnosis of bladder
cancer from urine samples (17), and
for the diagnosis of cervical cancer from captured cell lysates
obtained by laser capture microdissection of tissue samples
(18). SELDI-TOF/MS has also been
used for identifying and monitoring biomarkers of Alzheimer's
disease in cerebrospinal fluid (19),
and for analyzing saliva samples of patients with Sjögren's
syndrome (SS) (20). Therefore,
SELDI-TOF/MS technology may aid the identification of potential
prognostic or predictive markers for oral squamous cell carcinoma
(OSCC).
SELDI technique
The ProteinChip® (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) biology system used by SELDI-TOF/MS enables the
retention of proteins on a solid-phase chromatographic surface
prior to their subsequent ionization and detection by TOF/MS
(21). The chromatographic surfaces
in the various ProteinChip® arrays commercially available are
uniquely designed to retain proteins from a complex sample mixture
according to general or specific properties such as hydrophobicity
and charge (21). Each spot in the
array contains a chemically treated surface, including anionic,
cationic, hydrophobic, hydrophilic and metal surfaces (21). In the case of ionic exchange
ProteinChip® arrays, the operating mechanism is the reversible
binding of charged molecules to the surface of the chip (21). For analysis of the expression of
multiple samples simultaneously, 12×8-spot chips assembled in
96-well bioprocessors have been developed (21). The fraction of the proteome bound to
the chips may be subsequently analyzed by MS on the same chip,
resulting in a pattern of proteins characterized by their
mass-to-charge ratio (m/z) (22).
Currently, four types of chips exist: i) CM10
(Bio-Rad Laboratories, Inc.), which is a weak cation exchanger
whose surface has been negatively charged, thus enabling the
binding of proteins that are positively charged at a certain pH;
ii) Q10 (Bio-Rad Laboratories, Inc.), a strong anion exchanger with
quaternary ammonium groups positively charged on its surface, which
binds proteins that are negatively charged at alkaline conditions;
iii) IMAC30 (Bio-Rad Laboratories, Inc.), which binds proteins on
its surface by metal affinity; and iv) H50 (Bio-Rad Laboratories,
Inc.), which captures proteins through hydrophobic or reverse-phase
interaction (21).
The SELDI-TOF/MS instruments consist of three major
components, namely the ProteinChip® array, the reader and the
software. The ProteinChip® reader is an LDI-TOF/MS unit equipped
with a laser source. Upon activation of the laser, the sample
becomes irradiated, and is then subjected to DI, which liberates
gaseous ions from the ProteinChip® array (11). Next, these gaseous ions enter the
TOF/MS region of the instrument, which measures the m/z of each
protein based on its velocity through an ion chamber (11). Subsequent signal processing is
accomplished by a high-speed analog-to-digital converter linked to
a personal computer, whereby the detected proteins are displayed as
a series of peaks (11). The output
generated from the TOF/MS analysis of the samples is a trace
representing the relative abundance of the detected proteins vs.
their molecular weight (11). Thus,
the end result of a SELDI-TOF/MS analysis is a list of the
molecular weights of the detected proteins (11). Next, the proteomic profiles of all the
samples are analyzed by ProteinChip® Data Manager™ software version
3.5, in order to identify mass peaks (also known as clusters) that
are differentially expressed between two different groups (for
example, healthy vs. pathological samples).
SELDI technique and saliva
Sample collection and storage
The saliva samples for a SELDI-TOF/MS analysis must
be produced between 9:00 a.m. and 10:00 a.m. by prior mouth rinsing
with water (23). Donors must abstain
from eating, drinking, smoking or using oral hygiene products for
≥2 h prior to collection (24). The
saliva must be spitted directly into a clean 15 ml conical tube,
and a protease inhibitor cocktail must then be added to the samples
prior to further processing (24).
Subsequently, the saliva samples should be centrifuged for 50 min
at 13,000 × g, and the supernatants divided into aliquots and
frozen for storage at −80°C.
Reagents
Each chip must be pretreated with a specific binding
buffer, depending on its surface. Thus, 100 mM sodium acetate pH 4
must be used for CM10, while 100 mM Tris-HCl pH 8.8 and pH 7.4 must
be used for Q10 and IMAC30, respectively. The latter chip requires
preliminary loading with 0.1 M Cu2+, followed by
neutralization with 0.1 M sodium acetate (pH 4), prior to the
addition of 100 mM Tris-HCl (pH 7.4) as binding buffer. Milli-Q®
water (Merck Millipore, Darmstadt, Germany) may be used as
solvent.
Similarly, the sample must be diluted in the binding
buffer that is specific for the particular ProteinChip® tested, or
mixed (2:3 v/v) with a denaturing buffer solution such as DB3 (9 M
urea, 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
and 100 mM dithiothreitol; Sigma-Aldrich, St. Louis, MO, USA)
(3).
For all types of chips, 1 µl of matrix must be used,
which consists of 50% sinapinic acid (SPA) (Ciphergen Biosystems,
Inc., Fremont, CA, USA) solubilized in 50% acetonitrile (ACN)/0.5%
trifluoroacetic acid (TFA) (Sigma-Aldrich).
SELDI-TOF/MS analysis
Each chip must be pretreated at room temperature
with vigorous agitation with its specific binding buffer, according
to the manufacturer's protocol (Bio-Rad Laboratories, Inc.). Next,
the sample must be incubated on the chip in the presence of the
corresponding binding buffer for 30 min with continuous agitation
(3). Upon removing the sample from
the wells, the chip must then be washed three times with 150 µl
washing buffer (0.1 M sodium acetate [pH 4] for CM10; 0.1 M Tris
HCl [pH 8.8] for Q10; and 0.1 M Tris-HCl [pH 7.4] for IMAC30;
Sigma-Aldrich) while agitating, in order to remove the majority of
the salts present, which otherwise interfere with the subsequent MS
analysis (6).
A final wash should be performed with 200 µl
deionized Milli-Q® water, prior to air-dry the chip for 20 min
(3). Next, a saturated solution of
SPA must be prepared in 50% ACN/0.5% TFA, and diluted by 50% in
this solvent, in order to generate the matrix (3). Subsequently, 1 µl of this matrix must be
applied to each spot, and allowed to dry (25). The above procedure should be repeated
twice (25).
The matrix, also known as energy-absorbing matrix
(EAM), is a solution that crystallizes and promotes the ionization
of the proteins present in the sample once these have been dried on
the chip. Next, the chip must be placed in the SELDI ProteinChip®
reader for MS analysis (25).
Unless otherwise specified, it should be possible to
read all the chips by an automated protocol using the same
instrument conditions, namely, laser energy, 6,000 nJ; matrix
attenuation, 2,500 Da; focus mass, 10,000 Da; sample rate, 800 MHz;
coverage of the surface area of the spot, 25%; and acquired mass
range, 2,500-25,000 m/z) (24). All
the experiments must be performed in duplicate.
The proteomic profiles of all the samples must be
subsequently analyzed with ProteinChip® Data Manager™ software
version 3.5, in order to identify differentially expressed mass
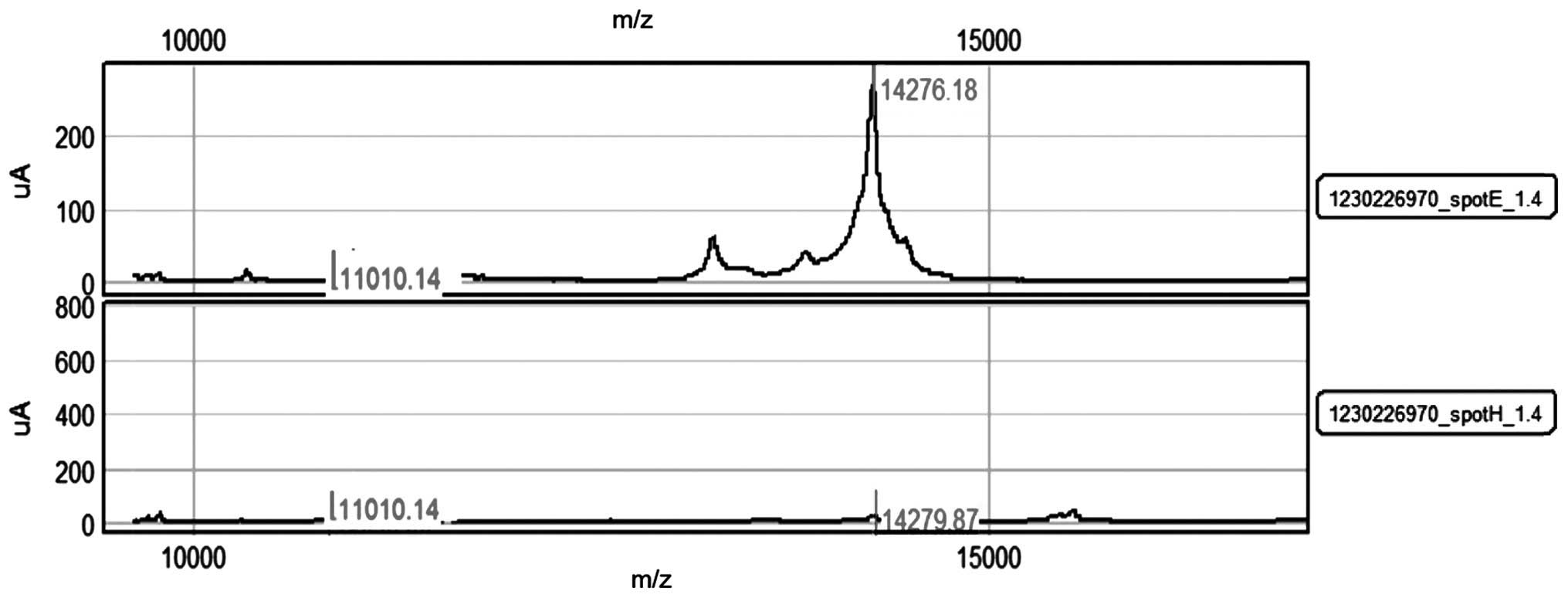
peaks (clusters) among groups of different samples (Fig. 1). These differences must be verified
by Mann-Whitney U test, whereby P<0.05 is considered to indicate
a statistically significant difference.
The analysis software employs classification and
regression tree, a multivariate analysis where the samples are
divided into two groups, termed the training and the testing set,
respectively. The intensity in µA units of all the mass peaks
identified in the training set is then used by Biomarker Pattern
Software (BPS)® (version 5.0; Ciphergen Biosystems, Inc.) to build
a classification tree.
The intensity of the peaks selected from the BPS®
analysis is regarded as a ‘root node’. Furthermore, it is possible
to develop an optimal threshold value to improve the stratification
of the two groups, based on the intensity of the mass peak measured
in each sample.
Discussion
The interest on saliva as diagnostic material has
markedly increased in recent years, since the information
associated with various substances present in this biological fluid
may aid the surveillance of general and oral health, and contribute
to the understanding of the pathogenesis of certain diseases
(26–30).
Proteins are the most important constituents of
saliva. Therefore, the full analysis and identification of the
human salivary proteome constitutes the first step towards the
identification of novel biomolecules associated with the salivary
state of health or disease (26–30).
As diagnostic material, saliva presents certain
disadvantages, including the influence exerted by the method of
collection and the degree of stimulation of the salivary flow on
the composition of the saliva samples (31,32). In
addition, saliva contains analytes at concentrations 1,000 times
lower than in plasma (33). Thus,
sensitive detection systems are required when using saliva as
diagnostic material (9).
SELDI-TOF/MS was originated by combining
MALDI-TOF/MS with surface chromatography, and has acquired an
important role in proteomic analysis in recent years (9). SELDI-TOF/MS is a highly efficient
technique that is particularly suitable for the study of small
peptides and proteins, since its fmol sensitivity complements the
results of 2D-PAGE analysis (9).
Previous studies have employed the SELDI technique
to search for potential biomarkers in the salivary proteome
(3,6,34–48) (Table
I).
 | Table I.Possible applications of
surface-enhanced laser desorption/ionization-time-of-flight/mass
spectrometry for clinical research on saliva. |
Table I.
Possible applications of
surface-enhanced laser desorption/ionization-time-of-flight/mass
spectrometry for clinical research on saliva.
| Reference | Disease | Proteins as
putative biomarkers in saliva |
|---|
| 34 | Salivary gland
secretion of Hirudo medicinalis | Identification of
45 compounds of 1,964–66.5 kDa |
| 35 | Breast cancer | Peaks at 18, 113,
170, 228 and 287 m/z |
| 20 | SS | Peaks at 11.8,
12.0, 14.3, 80.6 and 83.7 kDa were increased, while peaks at 17.3,
25.4 and 35.4 kDa were reduced in SS |
| 6 | HC | Improvement of
technology |
| 38 | HC | Improvement of
technology |
| 37 | Allo-HCT and
GVHD | Peaks at 11,760,
11,691, 11,946, 5,864, 15,149, 18,711, 17,556, 11,041, 94,737,
11,524, 38,788, 13,386, 80,197 and 27,885 m/z were increased
post-HCT |
| 36 | HC | Salivary proteome
associated with satiety and body mass index |
| 3 | HC | Improvement of
technology |
| 39 | HC | Improvement of
technology |
| 40 | OSCC and OLK | Identification of
peaks at 5,797, 2,902, 3,883 and 4,951 m/z in OSCC, and peaks at
5,818, 4,617 and 3,884 m/z in OLK |
| 41 | OSCC | Peak at 1,400 m/z
identified as truncated cystatin SA-I |
| 42 | OSCC and OLK | Peaks at 3,738 and
11,366 m/z were differentially expressed in OSCC vs. OLK |
| 43 | Fibromyalgia | Detection of
RhoGDI2 and calgranulin A and C |
| 47 | Orthodontics | Modifications of
the saliva proteome |
| 44 | Periodontitis in
obese patients | Peaks at 66,000,
15,200 and 15,900 m/z, and at 3,492, 3,448, 3,492 and at 5,378 m/z,
were increased in obese patients with and without periodontitis,
respectively |
| 45 | DS II | Peaks at 12,679 Da
(IMAC30), 13,264 Da (CM10) and 3,822 Da (Q10) were increased, while
peaks at 35,125 Da (CM10) and 12,954 Da (Q10) were reduced in DS
II |
| 48 | HC | Influence of sodium
chloride and sucrose on the presence of proteins of 2–20 kDa in
whole saliva following stimulation with different tastants |
| 46 | PSS | Differential
expression of peaks at 7,149, 7,192, 13,517, 13,714, 16,547 and
24,059 m/z in patients with PSS vs. HC |
In 2004, Baskova et al (34) performed the first study on saliva
using the SELDI technique, and detected 45 individual compounds of
molecular masses ranging from 1,964 to 66.5 kDa in salivary gland
secretions of the medicinal leech Hirudo medicinalis.
In 2006, Ryu et al (20) used parotid saliva to identify the most
significant salivary biomarkers in SS using SELDI-TOF/MS. The
results revealed eight peaks of molecular weights in the range of
10–200 kDa whose levels were >2-fold altered in the SS group,
compared with the non-SS group (P<0.005). Thus, the levels of
the peaks at 11.8, 12.0, 14.3, 80.6 and 83.7 kDa increased, while
those at 17.3, 25.4 and 35.4 kDa reduced in SS samples (22).
Using SELDI, Streckfus et al (35) identified the presence of proteins at
18, 113, 170, 228 and 287 m/z in the saliva samples of women with
breast cancer, and hypothesized that these proteins may be
efficient salivary biomarkers of breast cancer.
In 2007, Harthoorn et al (36) studied the human saliva proteome in
regards to satiety and body mass index using SELDI-TOF/MS, since
this technique provides a valuable and non-invasive way of
profiling, which enables the characterisation of novel and
differentially expressed peptides and proteins that may be used as
biomarkers of satiety and overweight.
Also in that year, Imanguli et al (37) evaluated by SELDI-TOF/MS the
alterations that occur in salivary proteins following allogeneic
hematopoietic stem cell transplantation (allo-HCT) in 41 patients
undergoing allo-HCT. The authors detected significant increases and
reductions in the levels of multiple salivary proteins.
The studies by Schipper et al (6,38) in 2007
demonstrated the possibility of using the SELDI technique to
interrogate the salivary proteome. In addition, the authors
reported the procedures required for the correct treatment of
saliva samples, while Papale et al (3,24) reported
a protocol that improved the quality and reproducibility of
SELDI-TOF/MS analysis.
In 2008, Esser et al (39) used SELDI to study the salivary
proteome in healthy subjects, revealing the presence of 218
proteins, 84 of which were also present in plasma. Based on a
comparison with previous proteomics studies on whole saliva, the
authors also identified 83 novel salivary proteins.
In 2009, Sun and Ping (40) studied the salivary proteome by SELDI,
and identified novel potential salivary biomarkers that may aid the
early diagnosis of oral cancer and forecast the transformation from
oral leukoplakia (OLK) to oral cancer and metastasis. The study
identified a differentiated pattern between patients with OSCC and
healthy subjects consisting of four biomarker peaks of 5,797,
2,902, 3,883 and 4,951 m/z, with a sensitivity of 88.24% and a
specificity of 93.33%. The study also identified a differentiated
pattern between patients with OSCC and patients with OLK, which
consisted of three biomarker peaks of 5,818, 4,617 and 3,884 m/z,
with 100.00% sensitivity and specificity. In addition, the results
of the study revealed a differentiated pattern between patients
with OSCC and those with local metastatic oral cancer, which
consisted of two biomarker peaks of 55,809 and 5,383 m/z, with a
sensitivity of 94.12% and a specificity of 85.71%.
In 2010, Shintani et al (41) also studied the salivary proteome of
patients with OSCC using SELDI analysis, and detected 26 proteins
with significantly different expression levels in patients with
OSCC, compared with healthy controls. In particular, the authors
identified the presence of a truncated cystatin SA-I, characterized
by the deletion of three amino acids from its N-terminus, in the
saliva of patients with OSCC.
These results are supported by the findings of He
et al (42), who in 2011
identified by SELDI-TOF/MS technology differential proteomic
patterns in serum, saliva and tissue samples of patients with OSCC,
compared with patients with OLK.
Recently, several SELDI-TOF/MS studies on the saliva
proteome have identified potential biomarkers for fibromyalgia
(43), periodontitis in obese
patients (44), denture stomatitis
(45), primary SS (46) and post-transplant complications,
including infections and graft-versus-host disease (37).
Previous studies on the saliva proteome using SELDI
technology have evaluated the modifications affecting the oral
mucosa and bones in patients undergoing orthodontic treatment
(47). In addition, a previous study
employed SELDI-TOF/MS to investigate the influence of sodium
chloride and sucrose solutions on the presence of proteins of 2–20
kDa in whole saliva (48). The
results revealed that oral stimulation with different tastants
affects the composition of salivary proteins in a protein- and
stimuli-dependent way, regardless of the glands of origin (48).
In conclusion, SELDI technology combines the
precision of MALDI-TOF/MS proteomic analysis and the
high-throughput nature of protein array analysis (9). Furthermore, the analysis of saliva may
enable the screening of a large number of patients, since the
collection of saliva samples is easy, non-invasive, inexpensive and
reduces the risk of cross-infections, contrarily to that of blood
and serum samples (37). Therefore,
the analysis of the salivary proteome by SELDI-TOF/MS may aid to
identify prognostic or diagnostic biomarkers, complement the
results of 2D-PAGE analysis and confirm the findings of
MALDI-TOF/MS analysis.
References
|
1
|
Humphrey SP and Williamson RT: A review of
saliva: Normal composition, flow and function. J Prosthet Dent.
85:162–169. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pedersen AM, Bardow A, Jensen SB and
Nauntofte B: Saliva and gastrointestinal functions of taste,
mastication, swallowing and digestion. Oral Dis. 8:117–129. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Papale M, Pedicillo MC, Di Paolo S,
Thatcher BJ, Lo Muzio L, Bufo P, Rocchetti MT, Centra M, Ranieri E
and Gesualdo L: Saliva analysis by surface-enhanced laser
desorption/ionization time-of-flight mass spectrometry
(SELDI-TOF/MS): From sample collection to data analysis. Clin Chem
Lab Med. 46:89–99. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grundmann O, Mitchell GC and Limesand KH:
Sensitivity of salivary glands to radiation: From animal models to
therapies. J Dent Res. 88:894–903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Battino M, Ferreiro MS, Gallardo I, Newman
HN and Bullon P: The antioxidant capacity of saliva. J Clin
Periodontol. 29:189–194. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schipper R, Loof A, de Groot J, Harthoorn
L, Dransfield E and van Heerde W: SELDI-TOF-MS of saliva:
Methodology and pre-treatment effects. J Chromatogr B Analyt
Technol Biomed Life Sci. 847:45–53. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vercauteren FG, Bergeron JJ, Vandesande F,
Arckens L and Quirion R: Proteomic approaches in brain research and
neuropharmacology. Eur J Pharmacol. 500:385–398. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van den Bergh G and Arckens L: Recent
advances in 2D electrophoresis: An array of possibilities. Expert
Rev Proteomics. 2:243–252. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Tarawneh SK and Bencharit S:
Applications of surface-enhanced laser desorption/ionization
time-of-flight (SELDI-TOF) mass spectrometry in defining salivary
proteomic profiles. Open Dent J. 3:74–79. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bensmail H and Haoudi A: Postgenomics:
Proteomics and bioinformatics in cancer research. J Biomed
Biotechnol. 2003:217–230. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Seibert V, Wiesner A, Buschmann T and
Meuer J: Surface-enhanced laser desorption ionization
time-of-flight mass spectrometry (SELDI TOF-MS) and ProteinChip
technology in proteomics research. Pathol Res Pract. 200:83–94.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Srinivas PR, Srivastava S, Hanash S and
Wright GL Jr: Proteomics in early detection of cancer. Clin Chem.
47:1901–1911. 2001.PubMed/NCBI
|
|
13
|
Jacobs IJ and Menon U: Progress and
challenges in screening for early detection of ovarian cancer. Mol
Cell Proteomics. 3:355–366. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bertucci F, Birnbaum D and Goncalves A:
Proteomics of breast cancer: Principles and potential clinical
applications. Mol Cell Proteomics. 5:1772–1786. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuo SC, Gananadha S, Scarlett CJ, Gill A
and Smith RC: Sporadic pancreatic polypeptide secreting tumors
(PPomas) of the pancreas. World J Surg. 32:1815–1822. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hellström M, Lexander H, Franzén B and
Egevad L: Proteomics in prostate cancer research. Anal Quant Cytol
Histol. 29:32–40. 2007.PubMed/NCBI
|
|
17
|
Vlahou A, Schellhammer PF, Mendrinos S,
Patel K, Kondylis FI, Gong L, Nasim S and Wright GL Jr: Development
of a novel proteomic approach for the detection of transitional
cell carcinoma of the bladder in urine. Am J Pathol. 158:1491–1502.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu S, Denny P, Denny P, et al:
Differentially expressed protein markers in human submandibular and
sublingual secretions. Int J Oncol. 25:1423–1430. 2004.PubMed/NCBI
|
|
19
|
Frankfort SV, van Campen JP, Tulner LR and
Beijnen JH: Serum amyloid beta peptides in patients with dementia
and age-matched non-demented controls as detected by
surface-enhanced laser desorption ionisation-time of flight mass
spectrometry (SELDI-TOF MS). Curr Clin Pharmacol. 3:144–154. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ryu OH, Atkinson JC, Hoehn GT, Illei GG
and Hart TC: Identification of parotid salivary biomarkers in
Sjogren's syndrome by surface-enhanced laser desorption/ionization
time-of-flight mass spectrometry and two-dimensional difference gel
electrophoresis. Rheumatology (Oxford). 45:1077–1086. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Issaq HJ, Veenstra TD, Conrads TP and
Felschow D: The SELDI-TOF MS approach to proteomics: Protein
profiling and biomarker identification. Biochem Biophys Res Commun.
292:587–592. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Engwegen JY, Gast MC, Schellens JH and
Beijnen JH: Clinical proteomics: Searching for better tumour
markers with SELDI-TOF mass spectrometry. Trends Pharmacol Sci.
27:251–259. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Navazesh M: Methods for collecting saliva.
Ann N Y Acad Sci. 694:72–77. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Papale M, Pedicillo MC, Thatcher BJ, Di
Paolo S, Lo Muzio L, Bufo P, Rocchetti MT, Centra M, Ranieri E and
Gesualdo L: Urine profiling by SELDI-TOF/MS: Monitoring of the
critical steps in sample collection, handling and analysis. J
Chromatogr B Analyt Technol Biomed Life Sci. 856:205–213. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ardito F, Giuliani M, Perrone D,
Giannatempo G, Di Fede O, Favia G, Campisi G, Colella G and Lo
Muzio L: Expression of salivary biomarkers in patients with Oral
Mucositis: Evaluation by SELDI-TOF/MS. Oral Dis. Nov 27–2015.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Denny P, Ho CM, Montemagno C, Shi W,
Qi F, Wu B, Wolinsky L and Wong DT: The oral fluid MEMS/NEMS chip
(OFMNC): Diagnostic and translational applications. Adv Dent Res.
18:3–5. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tabak LA: A revolution in biomedical
assessment: The development of salivary diagnostics. J Dent Educ.
65:1335–1339. 2001.PubMed/NCBI
|
|
28
|
Malamud D, Bau H, Niedbala S and Corstjens
P: Point detection of pathogens in oral samples. Adv Dent Res.
18:12–16. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Smoot LM, Smoot JC, Smidt H, Noble PA,
Könneke M, McMurry ZA and Stahl DA: DNA microarrays as salivary
diagnostic tools for characterizing the oral cavity's microbial
community. Adv Dent Res. 18:6–11. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Streckfus C and Bigler L: The use of
soluble, salivary c-erbB-2 for the detection and post-operative
follow-up of breast cancer in women: The results of a five-year
translational research study. Adv Dent Res. 18:17–24. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hofman LF: Human saliva as a diagnostic
specimen. J Nutr. 131:1621S–1625S. 2001.PubMed/NCBI
|
|
32
|
Kaufman E and Lamster IB: The diagnostic
applications of saliva - a review. Crit Rev Oral Biol Med.
13:197–212. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Christodoulides N, Mohanty S, Miller CS,
Langub MC, Floriano PN, Dharshan P, Ali MF, Bernard B, Romanovicz
D, Anslyn E, et al: Application of microchip assay system for the
measurement of C-reactive protein in human saliva. Lab Chip.
5:261–269. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baskova IP, Zavalova LL, Basanova AV,
Moshkovskii SA and Zgoda VG: Protein profiling of the medicinal
leech salivary gland secretion by proteomic analytical methods.
Biochemistry (Mosc). 69:770–775. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Streckfus CF, Bigler LR and Zwick M: The
use of surface-enhanced laser desorption/ionization time-of-flight
mass spectrometry to detect putative breast cancer markers in
saliva: A feasibility study. J Oral Pathol Med. 35:292–300. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Harthoorn LF, Schipper RG, Loof A,
Vereijken PF, Van Heerde WL and Dransfield E: Salivary biomarkers
associated with perceived satiety and body mass in humans.
Proteomics Clin Appl. 1:1637–1650. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Imanguli MM, Atkinson JC, Harvey KE, Hoehn
GT, Ryu OH, Wu T, Kingman A, Barrett AJ, Bishop MR, Childs RW, et
al: Changes in salivary proteome following allogeneic hematopoietic
stem cell transplantation. Exp Hematol. 35:184–192. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schipper R, Loof A, de Groot J, Harthoorn
L, van Heerde W and Dransfield E: Salivary protein/peptide
profiling with SELDI-TOF-MS. Ann N Y Acad Sci. 1098:498–503. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Esser D, Alvarez-Llamas G, de Vries MP,
Weening D, Vonk RJ and Roelofsen H: Sample stability and protein
composition of saliva: Implications for its use as a diagnostic
fluid. Biomark Insights. 3:25–27. 2008.PubMed/NCBI
|
|
40
|
Sun G and Ping FY: Application of saliva
protein fingerprints in the diagnosis of oral squamous cell cancer
by surface enhanced laser desorption ionization time of flight
mass. Zhonghua Kou Qiang Yi Xue Za Zhi. 44:664–667. 2009.(In
Chinese). PubMed/NCBI
|
|
41
|
Shintani S, Hamakawa H, Ueyama Y, Hatori M
and Toyoshima T: Identification of a truncated cystatin SA-I as a
saliva biomarker for oral squamous cell carcinoma using the SELDI
ProteinChip platform. Int J Oral Maxillofac Surg. 39:68–74. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
He H, Sun G, Ping F and Cong Y: A new and
preliminary three-dimensional perspective: Proteomes of
optimization between OSCC and OLK. Artif Cells Blood Substit
Immobil Biotechnol. 39:26–30. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Giacomelli C, Bazzichi L, Giusti L,
Ciregia F, Baldini C, Da Valle Y, De Feo F, Sernissi F, Rossi A,
Bombardieri S and Lucacchini A: MALDI-TOF and SELDI-TOF analysis:
‘Tandem’ techniques to identify potential biomarker in
fibromyalgia. Reumatismo. 63:165–170. 2011.(In Italian). View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rangé H, Léger T, Huchon C, Ciangura C,
Diallo D, Poitou C, Meilhac O, Bouchard P and Chaussain C: Salivary
proteome modifications associated with periodontitis in obese
patients. J Clin Periodontol. 39:799–806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bencharit S, Altarawneh SK, Baxter SS,
Carlson J, Ross GF, Border MB, Mack CR, Byrd WC, Dibble CF, Barros
S, et al: Elucidating role of salivary proteins in denture
stomatitis using a proteomic approach. Mol Biosyst. 8:3216–3223.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gallo A, Martini D, Sernissi F, Giacomelli
C, Pepe P, Rossi C, Riveros P, Mosca M, Alevizos I and Baldini C:
Gross cystic disease fluid protein-15
(GCDFP-15)/prolactin-inducible protein (PIP) as functional salivary
biomarker for primary Sjögren's syndrome. J Genet Syndr Gene Ther.
4:2013.PubMed/NCBI
|
|
47
|
Ciavarella D, Mastrovincenzo M, D'Onofrio
V, Chimenti C, Parziale V, Barbato E and Lo Muzio L: Saliva
analysis by surface-enhanced laser desorption/ionization
time-of-flight mass spectrometry (SELDI-TOF-MS) in orthodontic
treatment: First pilot study. Prog Orthod. 12:126–131. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Silletti E, Bult JH and Stieger M: Effect
of NaCl and sucrose tastants on protein composition of oral fluid
analysed by SELDI-TOF-MS. Arch Oral Biol. 57:1200–1210. 2012.
View Article : Google Scholar : PubMed/NCBI
|