Introduction
Ovarian cancer is the sixth most common type of
cancer diagnosed in women worldwide, accounting for ~4% of all
types of female cancer, and is the second most common gynecological
cancer, with ~75% of patients presenting with late-stage disease
(1–3).
Patients with ovarian cancer are generally treated with first-line
chemotherapy, including paclitaxel and platinum-based drugs
(4). Cisplatin, the first-generation
platinum-based drug, was gradually replaced by carboplatin and
oxaliplatin, which are second- and third-generation platinum-based
drugs, respectively (5,6). Cisplatin was replaced due to liver and
kidney toxicity, in addition to side effects such as mucositis,
neutropenia and alopecia. Since the genetic profiles of patients
with cancer have been associated with drug metabolism, patients may
receive targeted chemotherapy, which avoids unnecessary toxicity
(7). Pharmacogenetics is important in
cancer chemotherapy, and the prognosis of patients with cancer may
be explained according to their genetic background (8). The effects of platinum-based drugs are
controlled by genes involved in detoxification, including
glutathione S-transferase (GST)P1 and GSTM1 (9).
GST belongs to the multifunctional poly-2 protein
family, and is primarily responsible for catalyzing the interaction
between reduced glutathione hormone and electrophilic substances
(10). GST is important in protecting
tissues from damage caused by oxidative stress (11). The GSTM1 gene has two variants, and
encodes an enzyme that participates in the metabolism of
carcinogens (12).
The influence of genetic polymorphisms in the
different response to platinum-based chemotherapy has been observed
in certain respiratory and digestive types of cancer, including
non-small cell lung and colorectal cancer (13–17).
However, to the best of our knowledge, genetic polymorphisms in
patients with ovarian cancer who have been treated with paclitaxel
and platinum-based therapies have not been characterized thus far.
Therefore, the present study aimed to investigate the association
between genetic polymorphisms in drug metabolism and the overall
survival (OS) of patients with ovarian cancer treated with
paclitaxel and carboplatin-based chemotherapy. The present study
evaluated specific polymorphic genes, namely GSTP1 Ile105Val and
GSTM1 null, in order to assess the association between the
phenotype pattern and the OS of chemotherapy-treated patients with
ovarian serous cystadenocarcinoma.
Materials and methods
Patients and tissue samples
Tissue samples from 95 patients with ovarian serous
cystadenocarcinoma (median age, 53.6±11.5 years) were obtained from
Linyi People's Hospital (Linyi, China). All the patients were
diagnosed and treated at the Linyi People's Hospital between August
2005 and July 2013. The tissue samples were obtained during
diagnostic or therapeutic surgery. The patients were followed-up
once every 2–4 months for 2 years and then every 3–6 months in the
subsequent 3 years. Patients required follow-up once a year from 5
years onwards. The median follow-up of patients that were alive at
the end of the present study was 25.6±14.3 months (range, 3.0–92.0
months) All the patients provided written informed consent for the
use of tissues samples and participation in the study. The present
study was approved by the ethics committee at Linyi People's
Hospital.
Data from 104 patients was collected for the present
study, of which 9 patients were excluded. Patients that lacked
information regarding cancer stage or start dates of
chemotherapeutic treatment were excluded from the analysis. In
addition, patients with a survival time of <1 month were
excluded from the study. Information regarding the chemotherapeutic
treatment regimen was available for all the patients included in
the study. If the patients that possessed stage II–IV cancer, who
were treated with 2 cycles of paclitaxel and carboplatin-based
neoadjuvant chemotherapy (TP regimen) and 4–6 cycles of
chemotherapy post-surgery, were allergic to paclitaxel, their
levels of cancer antigen 125 were elevated following 1–2
chemotherapy cycles or computed tomography revealed that the volume
of the tumor was not reduced, then the patients would be
administered a different chemotherapy regimen, namely gemcitabine
(1,000 mg/m2, on days 1 and 8; treatment cycle, 21–28
days), ifosfamide (1,700 mg/m2, on days 1–3; treatment
cycle, 21–28 days) or topotecan (0.75 mg/m2, on days
1–5; treatment cycle, 21–28 days). In total, 95 patients that
received adjuvant or palliative chemotherapy, of which 56 patients
received TP regimen and 39 patients received alternative
chemotherapy, were eligible for analysis. The patients that
received the TP regimen were intravenously administered with
135–175 mg/m2 paclitaxel and 300–400 mg/m2
carboplatin on days 1 and 2, respectively. The treatment cycle was
21–28 days. The characteristics of the patients are presented in
Table I.
 | Table I.Characteristics of 95 patients with
ovarian serous cystadenocarcinoma. |
Table I.
Characteristics of 95 patients with
ovarian serous cystadenocarcinoma.
| Characteristic | Value |
|---|
| Age, years |
|
|
Median | 53.6 |
|
Range | 20.0–80.0 |
| Follow-up time,
months |
|
|
Median | 25.6 |
|
Range | 3.0–92.0 |
| Chemotherapy, n
(%) |
|
|
Paclitaxel + carboplatin | 56 (58.9) |
|
Other | 39 (41.1) |
| Tumor stage, n
(%) |
|
| II | 11 (11.6) |
| III | 63 (66.3) |
| IV | 21 (22.1) |
Genotyping of GST
The tumor tissues were obtained from Department of
Pathology, Linyi People's Hospital, where the pathologists used the
following procedure: Following the surgery, the tumor specimens
were immediately fixed with 10% formalin (Xilong Chemical Co.,
Ltd., Shantou, China) for 24 h. Subsequent to rinsing with running
tap water, the tissues were dehydrated with 70, 80 and 95% ethanol,
followed by 100% ethanol, which was changed 3 times. Subsequent to
clearing with xylene (twice), the tissues were immersed in paraffin
3 times (Leica, Wetzlar, Germany). The paraffin-embedded tissue
blocks were cut into 10-µm thick sections, from which genomic DNA
was extracted using the Wizard® Genomic DNA Purification Kit
(Promega Corporation, Madison, WI, USA). The germline mutations of
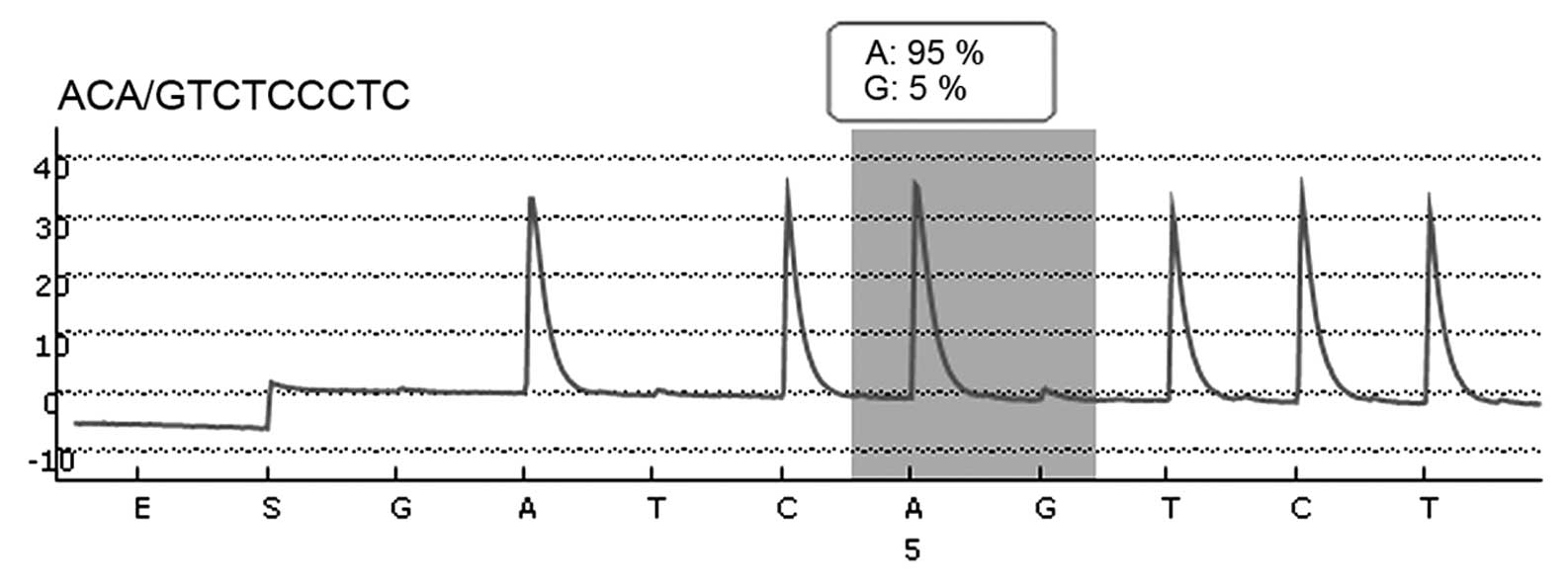
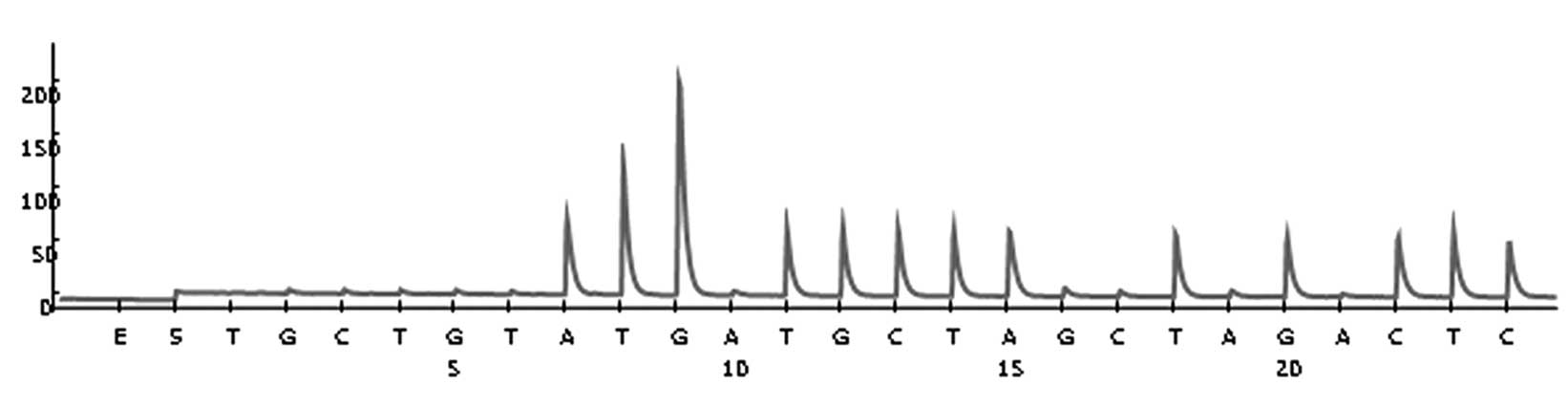
GSTP1 and GSTM1 were analyzed in all patients using pyrosequencing.
A total of four variants [three single nucleotide polymorphisms
(SNPs) and one gene deletion] were assessed using polymerase chain
reaction (PCR) and pyrosequencing, as previously described
(18).
The specific sequence primers (Shanghai Shengong
Biotechnology, Shanghai, China) used were as follows: GSTM1,
forward, 5′-CGCCATCTTGTGCTACATTGC-3′; reverse,
5′-CACAAATTCTGGATTGTAGCAGA-3′; and P (the reverse primer resulting
in a 237 bp fragment), 5′-GGCCTCCTCCTTGGCTGG-3′; GSTP1 Ile105Val,
forward, 5′-AATGACGGCGTGGAGGAC-3′; reverse,
5′-GGTCAGCCCAAGCCACCT-3′; and P (the reverse primer resulting in a
155 bp fragment), 5′-AGGACCTCCGCTGCAAAT-3′. The PCR cycle
conditions were as follows: Denaturation at 95°C for 5 min,
followed by 35 cycles of 95°C for 45 sec, 60°C for 30 sec and 72°C
for 30 sec. DNA amplification was performed using Taq DNA
Polymerase Master Mix RED (Biomol GmbH, Hamburg, Germany). For
quality control purposes and to verify the results, 10% of samples
were re-analyzed and 100% concordance was indicated (19). Negative control samples were included
in each amplification series. The presence of one or both GSTM1
alleles, as identified by the presence of a 237 bp fragment or
complete deletion (null genotype), was analyzed by electrophoresis
on a 1.2% agarose gel using DL500 as a DNA marker (Takara Bio,
Dalian, China). Electrophoresis was performed for 30 min at 120 V.
The gel image was captured using BDAdigital (Analytik Jena AG,
Jena, Germany). The absence of amplifiable GSTM1 (in the presence
of the GSTM4 co-amplified control) indicated a null genotype.
GSTM1 gene expression analysis by
reverse transcription-quantitative PCR (RT-qPCR)
Relative complementary DNA quantitation for GSTM1
and an internal reference gene (β-actin) was performed on
formalin-fixed, paraffin-embedded surgical specimens from 95
patients. Following standard tissue sample deparaffinization using
xylene and ethanol (20), samples
were lysed in a tris-chloride, ethylenediaminetetraacetic acid,
sodium dodecyl sulphate and proteinase K-containing buffer. RNA was
then extracted with phenol-chloroform-isoamyl alcohol followed by
precipitation with isopropanol in the presence of glycogen and
sodium acetate. RNA was resuspended in diethyl pyrocarbonate water
(Ambion, Inc.; Thermo Fisher Scientific) and treated with DNAseI
(Ambion, Inc.; Thermo Fisher Scientific) to avoid DNA
contamination. Complementary DNA was synthesized using the Moloney
Murine Leukemia Virus retrotranscriptase enzyme. Template cDNA was
added to Taqman Universal MasterMix (Applied Biosystems; Thermo
Fisher Scientific, Inc.) in a 12.5-µl reaction with specific primers
and probes for each gene. The primer and probe sets were designed
using PrimerExpress 2.0 Software (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and the RefSeq sequences (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene).
The sequences of the primers and probes used were as follows: GSTM1
forward, 5′-CCCAGAGCAACGCCATCT-3′; reverse,
5′-TCCACACGAATCTTCTCCTCTTC-3′; probe,
(FAM)-5′-CTACATTGCCCGCAAGCACAACCTG-3′-(TAMRA). β-actin (internal
reference gene) forward, 5′-TGAGCGCGGCTACAGCTT-3′; reverse,
5′-TCCTTAATGTCACGCACGATTT-3′; probe,
(FAM)-5′-ACCACCACGGCCGAGCGG-3′-(TAMRA). Quantification of gene
expression was carried out using the LineGene K system (Bioer
Technology Co., Ltd.).
Relative gene expression quantification was
calculated according to the comparative cycle threshold (Cq) method
using β-actin as an endogenous control Final results were
determined as follows: 2−ΔΔCq, where Δ Cq values of the
sample are determined by subtracting the Cq value of the target
gene from the value of the β-actin gene.
Statistical analysis
SPSS software version 17.0 (SPSS Inc., Chicago, IL,
USA) was used for statistical analysis. P<0.05 was considered to
indicate a statistically significant difference. All statistical
tests were two-sided with a significance level of α=0.05. Genotype
data were categorized as follows: Homozygous carriers of the
wild-type allele for GSTP1 polymorphism (Val105Val); carriers of
the heterozygous allele or possessing one gene copy (GSTP1
Ile105Val); and carriers of the homozygous variant allele or
possessing no copy (GSTP1 Ile105Ile). Patients with >1 copy of
GSTM1 were considered to exhibit the non-null genotype, whereas
patients possessing no copies of GSTM1 were considered to exhibit
the null genotype. The OS of patients was calculated as the time
between the start of the chemotherapeutic treatment and the date of
the last follow-up or the date when the patient succumbed to
disease. Kaplan-Meier survival function analysis was used to
evaluate the association between GST genotypes and the OS of
patients. The log-rank test was used to calculate the difference
between OS and genotype. Cox proportional hazards regression
analysis was used to calculate multivariable adjusted hazard ratios
(HRs) and their corresponding 95% confidence interval (CI).
Results
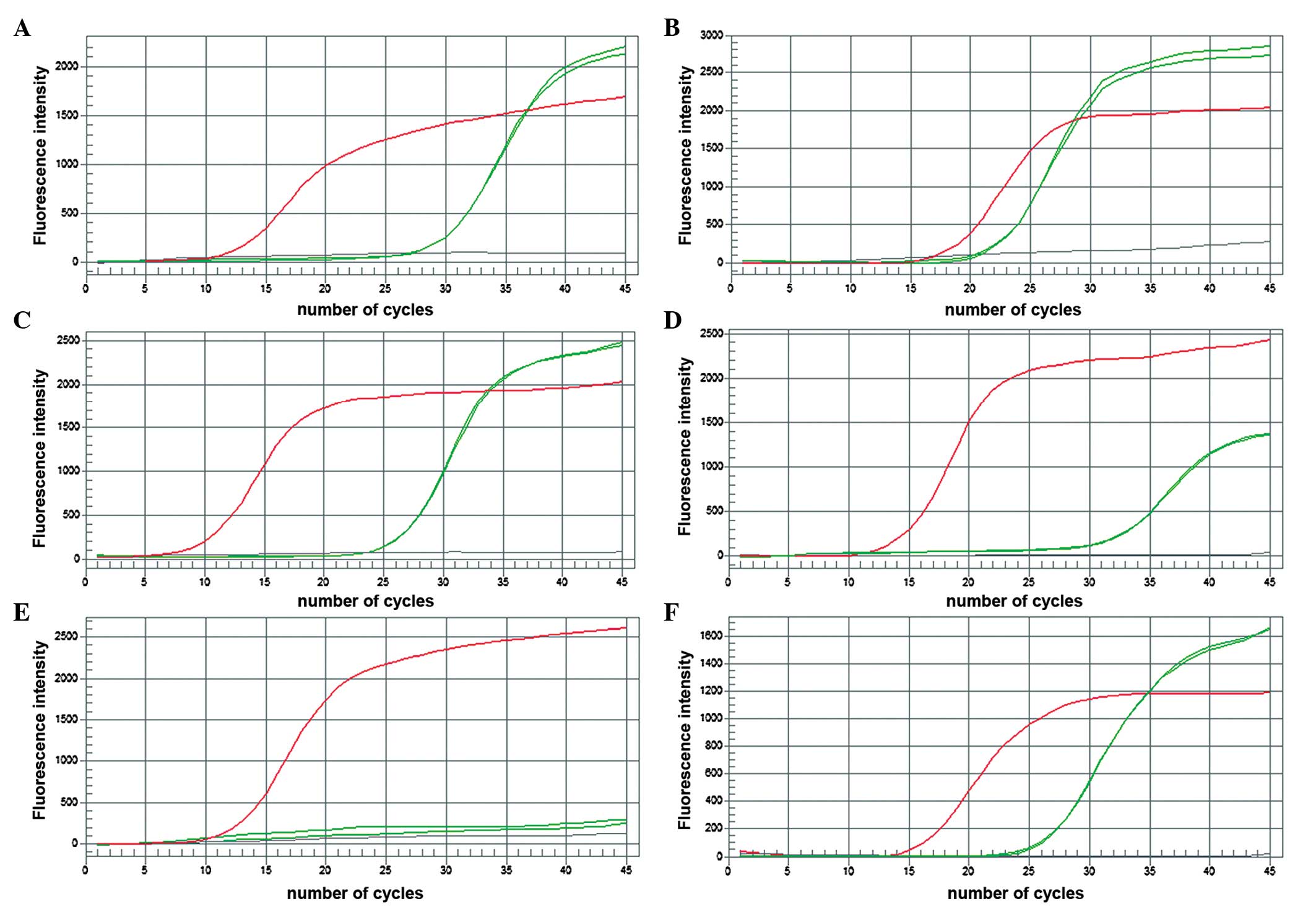
The frequency of GSTM1 positive and negative
individuals was determined by genotyping using multiplex-PCR as
shown in Fig. 1. Relative GSTM1 gene
expression was also quantified (Fig.
2).
OS of patients with ovarian serous
cystadenocarcinoma
Patients that received TP or an alternative form of
chemotherapy were followed-up for a median time of 25.6±14.3
months. Kaplan-Meier survival analysis demonstrated that the OS of
patients did not differ significantly between GSTP1 genotypes
(log-rank test, P=0.17). Cox proportional hazards regression
analysis revealed that, since the start of treatment, there was not
a significant association between the GSTP1 isoleucine allele and
OS for heterozygous carriers of the isoleucine allele (HR, 1.78;
95% CI, 0.77–4.12; P=0.18) and no homozygous carriers of the valine
allele had been detected (HR, 0.00).
Kaplan-Meier survival analysis also revealed that
the OS did not differ significantly between GSTM1 genotypes
(log-rank test, P=0.83). Compared with carriers of two copies of
GSTM1, patients with ≤1 copies of GSTM1 exhibited no decrease in
the risk of mortality following chemotherapy [HR, 0.96, 95% CI,
0.37–2.51 (P=0.94) for patients with one copy of GSTM1 vs. HR,
0.71, 95% CI, 0.22–2.28 (P=0.56) for patients with no copies of
GSTM1, respectively] (Table II;
Figs. 3–6).
 | Table II.HR of the overall survival of 95
patients with ovarian serous cystadenocarcinoma, according to their
GSTP1 and GSTM1 polymorphisms. |
Table II.
HR of the overall survival of 95
patients with ovarian serous cystadenocarcinoma, according to their
GSTP1 and GSTM1 polymorphisms.
| Genetic
polymorphism | Patients, n | Mortalities, n | P-valuea | HR | 95% CI | P-valueb |
|---|
| GSTP1 |
|
|
|
|
|
|
| A/A | 37 | 11 | – | 1.00 | – | – |
| A/G | 58 | 11 | 0.17 | 1.78 | 0.77–4.12 | 0.18 |
| G/G | 0 | 0 | 0.00 | 0.00 | 0.00–0.00 | 0.00 |
| GSTM1 |
|
|
|
|
|
|
|
Homozygous (2 copies) | 27 | 8 | – | 1.00 | – | – |
|
Heterozygous (1 copy) | 20 | 4 | – | 0.96 | 0.37–2.51 | 0.94 |
|
Homozygous (0 copies) | 48 | 10 | 0.83 | 0.71 | 0.22–2.28 | 0.56 |
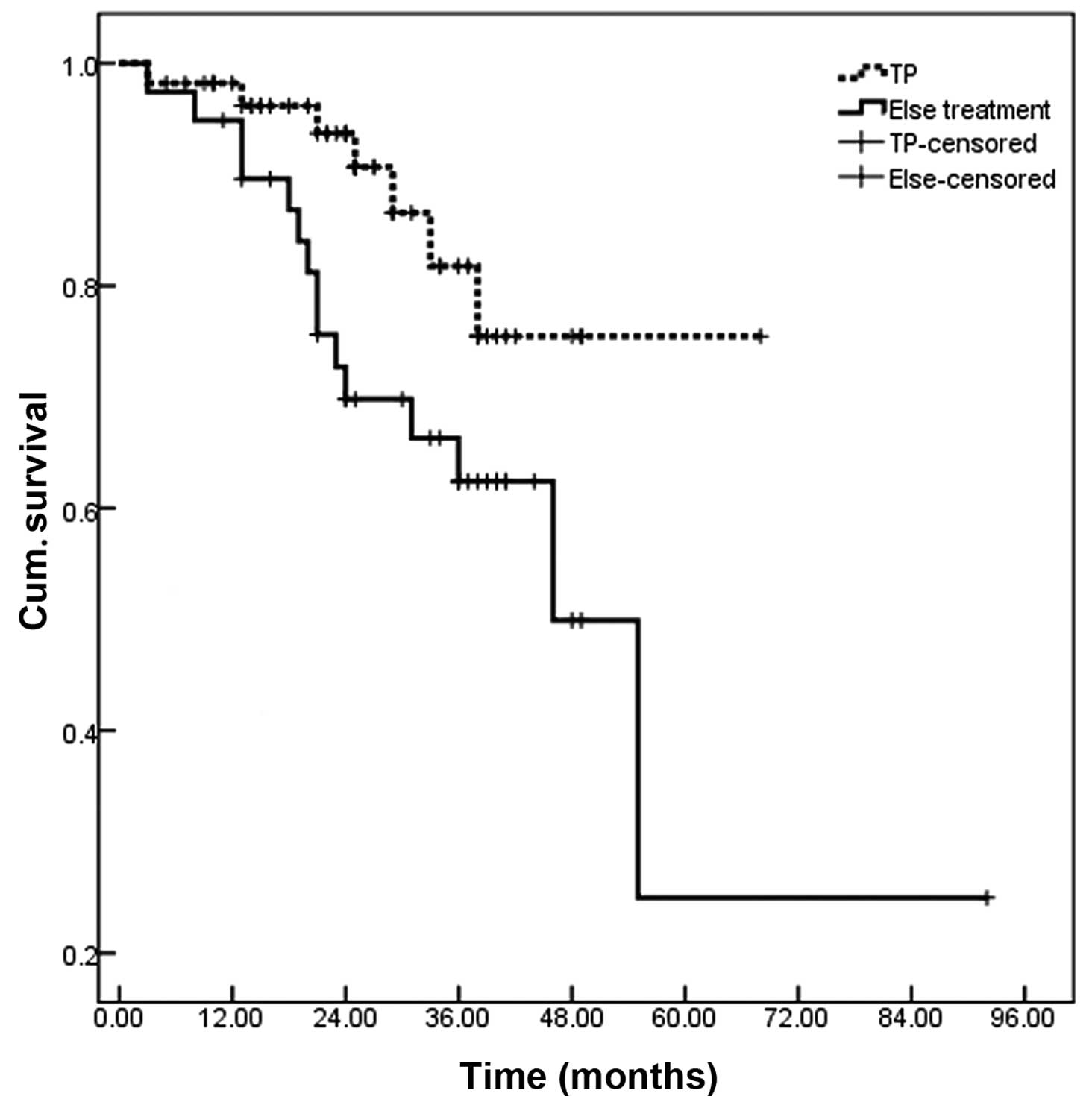
Overall, there were no associations between
polymorphisms in GSTP1 and GSTM1 and the OS of patients following
adjuvant chemotherapy. However, the OS of patients administered
with TP was significantly increased, compared with patients that
received other type of chemotherapy (P=0.035; Fig. 7).
Discussion
Platinum-based chemotherapy drugs are commonly used
to treat certain solid tumors, including non-small cell lung and
colorectal cancer, in order to induce the formation of DNA adducts,
which contributes to the death of tumor cells (21). However, DNA repair and drug metabolism
may hinder the prognosis of patients that are administered
platinum-based chemotherapy (22,23).
Variations in the GSTP1 and GSTM1 genes have been
extensively studied, due to their capacity to modulate the drug
response in patients with cancer (24). GSTM1 deficiency has been reported to
increase the risk of developing head-neck tumors, squamous cell
carcinoma, and lung, colorectal, bladder and breast cancer
(25–27).
The present study aimed to evaluate the association
between genetic polymorphisms in the GSTP1 and GSTM1 genes and the
OS of patients with ovarian serous cystadenocarcinoma treated with
chemotherapy. GSTP1 is a member of the GST superfamily, and is
important in the defense function of cells (28). GSTP1 Ile105Val polymorphism leads to a
decreased ability in cell defense, thus increasing the sensitivity
of an individual to platinum-based chemotherapy (29,30).
However, the results of the Cox proportional hazards regression in
the present study revealed no significant association between the
GSTP1 Ile105Val isoleucine allele, and no homozygous carriers of
the valine allele were detected. None of the polymorphisms in the
GSTP1 and GSTM1 genes that were evaluated in the present study were
observed to be significantly associated with the OS of patients
that received chemotherapy TP regimen or an alternative
chemotherapeutic drug. Similarly, a previous study conducted on 65
patients with colorectal cancer that received first-line
chemotherapy of oxaliplatin did not identify an association between
GSTP1 genotype and survival (16).
However, a dose-dependent association between the number of GSTP1
valine alleles and survival in certain patients with cancer that
received second-line oxaliplatin treatment was reported by a
previous study, although no association between GSTM1 genotype and
survival was observed (31). The
present results revealed that there was not a significant
association between the GSTP1 isoleucine allele and the risk of
mortality for heterozygous carriers of the isoleucine allele (HR,
1.78; 95% CI, 0.77–4.12; P=0.18) and homozygous carriers of the
valine allele (HR, 0.00; no homozygous carriers of the valine
allele were detected) using Cox proportional hazards regression
analysis.
Townsend and Tew (32)
reported that GSTM1 is important in the detoxification of various
carcinogens, and is associated with the metabolism of various
chemotherapeutic agents. The results of the present study revealed
that the OS of patients did not vary significantly according to
their GSTM1 genotype. Patients with ≤1 copies of GSTM1 exhibited no
decreased risk of mortality following chemotherapy, compared with
patients with two copies of GSTM1.
In conclusion, the present study demonstrates that
there is a decreased risk of mortality for chemotherapy-treated
patients that have a reduced copy number of the GSTM1 allele. In
addition, patients that were GSTP1 heterozygous carriers of the
valine allele (A/G) had an increased risk of mortality, which is
contrary to the results of other studies (33,34). Tumor
characteristics, including classification and stage, may have
resulted in the inconsistent results observed. Additional studies
with larger sample sizes are required to confirm the results of the
present study.
Acknowledgements
The present study was supported by the Medical and
Health Science Technology Development Project of Shandong Province,
China (grant no. 2015WS0377) and the Natural Science Foundation of
Shandong Province, China (grant no. ZR2010CQ035).
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Partridge EE and Barnes MN: Epithelial
ovarian cancer: Prevention, diagnosis, and treatment. CA Cancer J
Clin. 49:297–320. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harries M and Gore M: Part. I Chemotherapy
for epithelial ovarian cancer-treatment at first diagnosis. Lancet
Oncol. 3:529–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Petrelli F, Zaniboni A, Coinu A, Cabiddu
M, Ghilardi M, Sgroi G and Barni S: Cisplatin or not in advanced
gastric cancer: A systematic review and meta-analysis. PLoS One.
8:e830222013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tan S, Peng X, Peng W, Zhao Y and Wei Y:
Enhancement of oxaliplatin-induced cell apoptosis and tumor
suppression by 3-methyladenine in colon cancer. Oncol Lett.
9:2056–2062. 2015.PubMed/NCBI
|
|
7
|
Kweekel DM, Gelderblom H and Guchelaar HJ:
Pharmacology of oxaliplatin and the use of pharmacogenomics to
individualize therapy. Cancer Treat Rev. 31:90–105. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jun L, Haiping Z and Beibei Y: Genetic
polymorphisms of GSTP1 related to response to
5-FU-oxaliplatin-based chemotherapy and clinical outcome in
advanced colorectal cancer patients. Swiss Med Wkly. 139:724–728.
2009.PubMed/NCBI
|
|
9
|
Shim HJ, Yun JY, Hwang JE, Bae WK, Cho SH,
Lee JH, Kim HN, Shin MH, Kweon SS, Lee JH, et al: BRCA1 and XRCC1
polymorphisms associated with survival in advanced gastric cancer
treated with taxane and cisplatin. Cancer Sci. 101:1247–1254. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan H, Jia H, Gao H, Guo X and Xu B:
Identification, genomic organization, and oxidative stress response
of a sigma class glutathione S-transferase gene (AccGSTS1) in the
honey bee, Apis cerana cerana. Cell Stress Chaperones.
18:415–426. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vontas JG, Small GJ and Hemingway J:
Glutathione S-transferases as antioxidant defence agents confer
pyrethroid resistance in Nilaparvata lugens. Biochem J.
357:65–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hayes JD and Pulford DJ: The glutathione
S-transferase supergene family: Regulation of GST and the
contribution of the isoenzymes to cancer chemoprotection and drug
resistance. Crit Rev Biochem Mol Biol. 30:445–600. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chan EC, Lam SY, Fu KH and Kwong YL:
Polymorphisms of the GSTM1, GSTP1, MPO, XRCC1 and NQO1 genes in
Chinese patients with non-small cell lung cancers: Relationship
with aberrant promoter methylation of the CDKN2A and RARB genes.
Cancer Genet Cytogenet. 162:10–20. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye F, Liu Z, Tan A, Liao M, Mo Z and Yang
X: XRCC1 and GSTP1 polymorphisms and prognosis of oxaliplatin-based
chemotherapy in colorectal cancer: A meta-analysis. Cancer
Chemother Pharmacol. 71:733–740. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Economopoulos KP and Sergentanis TN:
GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: A
comprehensive meta-analysis. Eur J Cancer. 46:1617–1631. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Funke S, Timofeeva M, Risch A, Hoffmeister
M, Stegmaier C, Seiler CM, Brenner H and Chang-Claude J: Genetic
polymorphisms in GST genes and survival of colorectal cancer
patients treated with chemotherapy. Pharmacogenomics. 11:33–41.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zaanan A, Dalban C, Emile JF, Blons H,
Fléjou JF, Goumard C, Istanbullu M, Calmel C, Alhazmi K, Validire
P, et al: ERCC1, XRCC1 and GSTP1 single nucleotide polymorphisms
and survival of patients with colon cancer receiving
oxaliplatin-based adjuvant chemotherapy. J Cancer. 5:425–432. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marsh S, King CR, Garsa AA and McLeod HL:
Pyrosequencing of clinically relevant polymorphisms. Methods Mol
Biol. 311:97–114. 2005.PubMed/NCBI
|
|
19
|
Butkiewicz D, Rusin M, Sikora B, Lach A
and Chorąży M: An association between DNA repair gene polymorphisms
and survival in patients with resected non-small cell lung cancer.
Mol Biol Rep. 38:5231–5241. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kizys MM, Cardoso MG, Lindsey SC, Harada
MY, Soares FA, Melo MC, Montoya MZ, Kasamatsu TS, Kunii IS,
Giannocco G, et al: Optimizing nucleic acid extraction from thyroid
fine-needle aspiration cells in stained slides,
formalin-fixed/paraffin-embedded tissues, and long-term stored
blood samples. Arq Bras Endocrinol Metabol. 56:618–626. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chai H, Pan J, Zhang X, Zhang X, Shen X,
Li H, Zhang K, Yang C, Sheng H and Gao H: ERCC1 C118T associates
with response to FOLFOX4 chemotherapy in colorectal cancer patients
in Han Chinese. Int J Clin Exp Med. 5:186–194. 2012.PubMed/NCBI
|
|
22
|
Chen YC, Tzeng CH, Chen PM, Lin JK, Lin
TC, Chen WS, Jiang JK, Wang HS and Wang WS: Influence of GSTP1
I105V polymorphism on cumulative neuropathy and outcome of FOLFOX-4
treatment in Asian patients with colorectal carcinoma. Cancer Sci.
101:530–535. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lv H, Li Q, Qiu W, Xiang J, Wei H, Liang
H, Sui A and Liang J: Genetic polymorphism of XRCC1 correlated with
response to oxaliplatin-based chemotherapy in advanced colorectal
cancer. Pathol Oncol Res. 18:1009–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ali-Osman F, Akande O, Antoun G, Mao JX
and Buolamwini J: Molecular cloning, characterization and
expression in Escherichia coli of full-length cDNAs of three
human glutathione S-transferase Pi gene variants. Evidence for
differential catalytic activity of the encoded proteins. J Biol
Chem. 272:10004–10012. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Godschalk RW, Dallinga JW, Wikman H, Risch
A, Kleinjans JC, Bartsch H and Van Schooten FJ: Modulation of DNA
and protein adducts in smokers by genetic polymorphisms in GSTM1,
GSTT1, NAT1 and NAT2. Pharmacogenetics. 11:389–398. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gertig DM, Stampfer M, Haiman C, Hennekens
CH, Kelsey K and Hunter DJ: Glutathione S-transferase GSTM1 and
GSTT1 polymorphisms and colorectal cancer risk: A prospective
study. Cancer Epidemiol Biomarkers Prev. 7:1001–1005.
1998.PubMed/NCBI
|
|
27
|
Slattery ML, Potter JD, Samowitz W, Bigler
J, Caan B and Leppert M: NAT2, GSTM-1, cigarette smoking, and risk
of colon cancer. Cancer Epidemiol Biomarkers Prev. 7:1079–1084.
1998.PubMed/NCBI
|
|
28
|
Li QF, Yao RY, Liu KW, Lv HY, Jiang T and
Liang J: Genetic polymorphism of GSTP1: Prediction of clinical
outcome to oxaliplatin/5-FU-based chemotherapy in advanced gastric
cancer. J Korean Med Sci. 25:846–852. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hong J, Han SW, Ham HS and Kim TY, Choi
IS, Kim BS, Oh DY, Im SA, Kang GH, Bang YJ and Kim TY: Phase II
study of biweekly S-1 and oxaliplatin combination chemotherapy in
metastatic colorectal cancer and pharmacogenetic analysis. Cancer
Chemother Pharmacol. 67:1323–1331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Watson MA, Stewart RK, Smith GB, Massey TE
and Bell DA: Human glutathione S-transferase P1 polymorphisms:
Relationship to lung tissue enzyme activity and population
frequency distribution. Carcinogenesis. 19:275–280. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stoehlmacher J, Park DJ, Zhang W, Groshen
S, Tsao-Wei DD, Yu MC and Lenz HJ: Association between glutathione
S-transferase P1, T1, and M1 genetic polymorphism and survival of
patients with metastatic colorectal cancer. J Natl Cancer Inst.
94:936–942. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Townsend DM and Tew KD: The role of
glutathione-S-transferase in anti-cancer drug resistance. Oncogene.
22:7369–7375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang MY, Huang ML, Chen MJ, Lu CY, Chen
CF, Tsai PC, Chuang SC, Hou MF, Lin SR and Wang JY: Multiple
genetic polymorphisms in the prediction of clinical outcome of
metastatic colorectal cancer patients treated with first-line
FOLFOX-4 chemotherapy. Pharmacogenet Genomics. 21:18–25. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suh KW, Kim JH, Kim DY, Kim YB, Lee C and
Choi S: Which gene is a dominant predictor of response during
FOLFOX chemotherapy for the treatment of metastatic colorectal
cancer, the MTHFR or XRCC1 gene? Ann Surg Oncol. 13:1379–1385.
2006. View Article : Google Scholar : PubMed/NCBI
|