Introduction
Mutations found in the nucleophosmin gene
(NPM1) are novel molecular abnormalities identified in acute
myeloid leukemia (AML). NPM1 mutations are present in ~60%
of cytogenetically normal AML cases, and they are an important
prognostic factor (1,2). Therefore, AML with mutated NPM1
has been defined as a provisional entity in the 4th edition of the
World Health Organization classification (3). The mutations typically appear as a 4
base pair (bp) insertion at position 956 through 971 in exon 12,
resulting in frame-shift mutations (1). To date, 61 known types of mutations have
been reported (2,4–7). The most
common NPM1 mutation is an insertion of TCTG at position
956–959, referred to as type A (see Table
I), and it is detected in ~80% of NPM1 mutation cases
(1,8,9).
 | Table I.Primers, probes, and sequences of the
mutated region. |
Table I.
Primers, probes, and sequences of the
mutated region.
| Variable | Sequence |
|---|
| PCR primers |
|
|
Forward |
5′-tgatgtctatgaagtgttgtggttcc-3′ |
|
Reverse |
5′-ctctgcattataaaaaggacagccag-3′ |
| Quenching probes |
|
| 1 |
acttcctccactgccagagatc-(BODIPY FL) |
| 2 | (BODIPY
FL)-cctccactgccagagatcttgaa-P |
| 3 | (BODIPY
FL)-ctattcaagatctctggcagt-P |
| Sequences |
|
|
Wild-type |
caggctattcaagatctctggcagtggagg |
| Type A
mutant |
caggctattcaagatctctgtctggcagtggagg |
Direct sequencing is the current standard method for
detecting NPM1 mutations. Unfortunately, this method is
costly and time consuming. In addition, the sensitivity of this
method is low, with the lower limit of detection beginning at ~20%
(10). Therefore, polymerase chain
reaction (PCR) methods such as electrophoresis (11), melting curve analysis (12), high-resolution melting (HRM) analysis
which detects the different melting temperatures of the PCR
products (13), locked nucleic acid
clamp-mediated PCR (10,14), and capillary electrophoresis (15,16) have
been investigated as potentially more sensitive methods for
detection of this 4 bp mutation.
Recently, the quenching probe (QP) method has been
developed as a novel technique for mutation detection (17,18). A QP
is an oligonucleotide with a fluorescent dye-modified cytosine at
the 5′ or 3′ end. After PCR amplification of the sequences
including the mutation site, a melting curve analysis using QPs is
performed. At low temperature, QPs hybridize with PCR products, and
their fluorescence is quenched by an electron transfer to adjacent
guanine bases in the PCR products. With an increase in temperature,
the QPs dissociate from the products, causing an increased
fluorescent signal emission (19).
Because QPs dissociate from unmatched products at lower
temperatures than perfectly matched products, it is possible to
detect mutant alleles (20). In the
present study, the sensitivity and effectiveness of a newly
established QP method was examined.
Materials and methods
Cells and DNA extraction
Two leukemia cell lines were used: an AML cell line,
OCI/AML3, with a heterozygous type A NPM1 mutation (9) and a megakaryoblastic leukemia cell line,
M-07e, with homozygous wild-type NPM1. DNA was extracted from the
cells by the SepaGene agglutination partition method (EIDIA, Tokyo,
Japan). To examine the sensitivity of the detection method, mixed
samples of OCI/AML3 DNA and M-07e DNA in various ratios were used.
Blood samples were obtained from 5 AML patients prior to treatment,
with their informed consent. The DNA extracted from these samples
was also examined. The study was approved by the Ethics Review
Board in Tokyo Medical and Dental University (Tokyo, Japan).
QP method
The QP method from a previously reported protocol
(18) was performed using a
LightCycler Nano™ (Roche Diagnostics, Mannheim, Germany). The
sequences of PCR primers and QPs are presented in Table I. A total of 3 QPs with different
sequences were designed to compare efficiency. The PCR cycling
conditions were as follows: A 95°C hold (10 min); 10 cycles at 95°C
(10 sec), 65–55°C ramp (10 sec, 1°C/step), 72°C (30 sec); followed
by 45 cycles of 95°C (10 sec), 55°C (10 sec), and 72°C (30 sec).
The reaction volume consisted of 19.5 µl with 18 ng DNA sample, 1.5
mM MgCl2, 0.2 µM primers, 0.2 µM QP, and 4 µl reaction
mix of LC 480 Genotyping Master (Roche Diagnostics). Upon
completion of the PCR cycles, the temperature was maintained at
95°C for 1 min, followed by 45°C for 1 min, and then gradually
increased to 95°C at a rate of 0.2°C/sec, during which the
fluorescent signal was continually acquired. The curves derived
from the fluorescent intensity with respect to temperature,
-dFluorescence/dTemperature (−dF/dT), were obtained using
LightCycler Nano™ software version 1.0 (Roche Diagnostics). Each
assay was performed in duplicate to verify the reproducibility of
the method.
Direct sequencing
For confirmation of sequences, the PCR products were
treated with Amicon Ultra Centrifugal Filter Units (Sigma-Aldrich,
St. Louis, MO, USA) to remove the primers and dNTPs, and then
sequenced using a 3130×1 Genetic Analyzer and BigDye terminator
Reaction kit v3.1 (Applied Biosystems, Foster city, CA, USA)
according to the manufacturer's protocol.
Results
Discrimination between wild-type and
mutant alleles
The curves for -dF/dT as determined by QP analysis
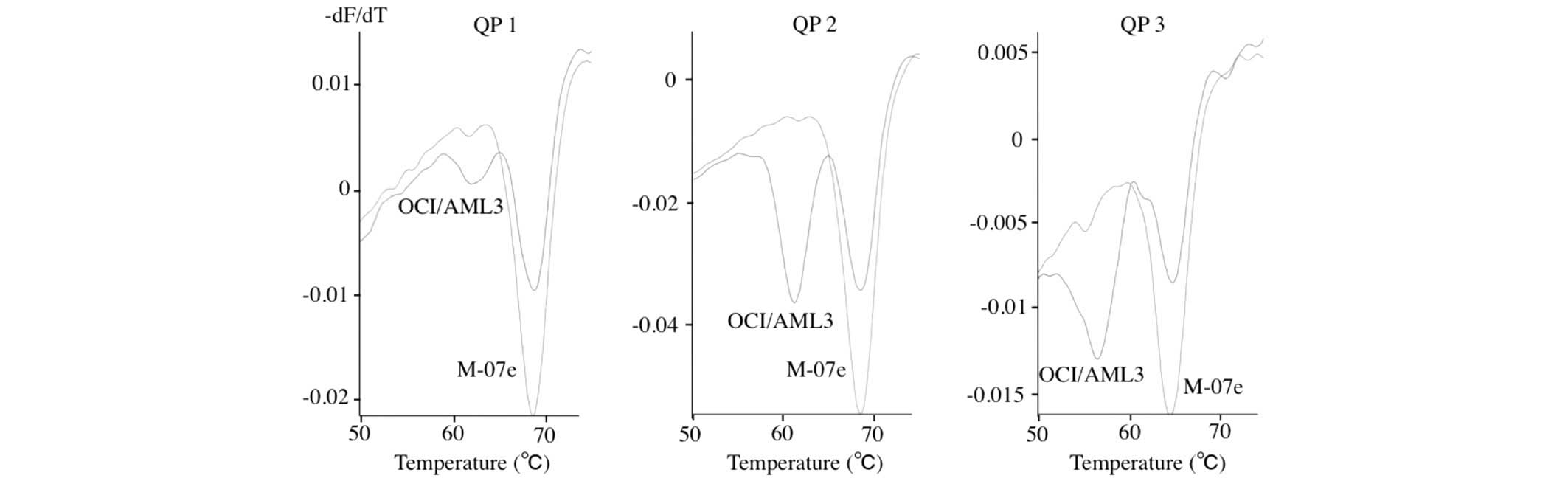
using the three variations of QPs (QP 1, QP 2, and QP 3) are shown
in Fig. 1. DNA from M-07e cells
possessing the homozygous wild-type alleles presented with single
concave-up curves, with the lowest point at about 65–68°C. DNA from
OCI/AML3 cells possessing the wild-type allele and the type A
mutant allele presented two concave-up curves with lowest points at
65–68°C and 55–61°C, respectively. In comparing the melting
profiles of the three variations of QPs investigated, QP 2 was the
most discriminative probe; the wild-type allele and type A mutant
allele presented curves with the lowest points at 68°C and 61°C,
respectively. Therefore, further QP analysis was performed using QP
2.
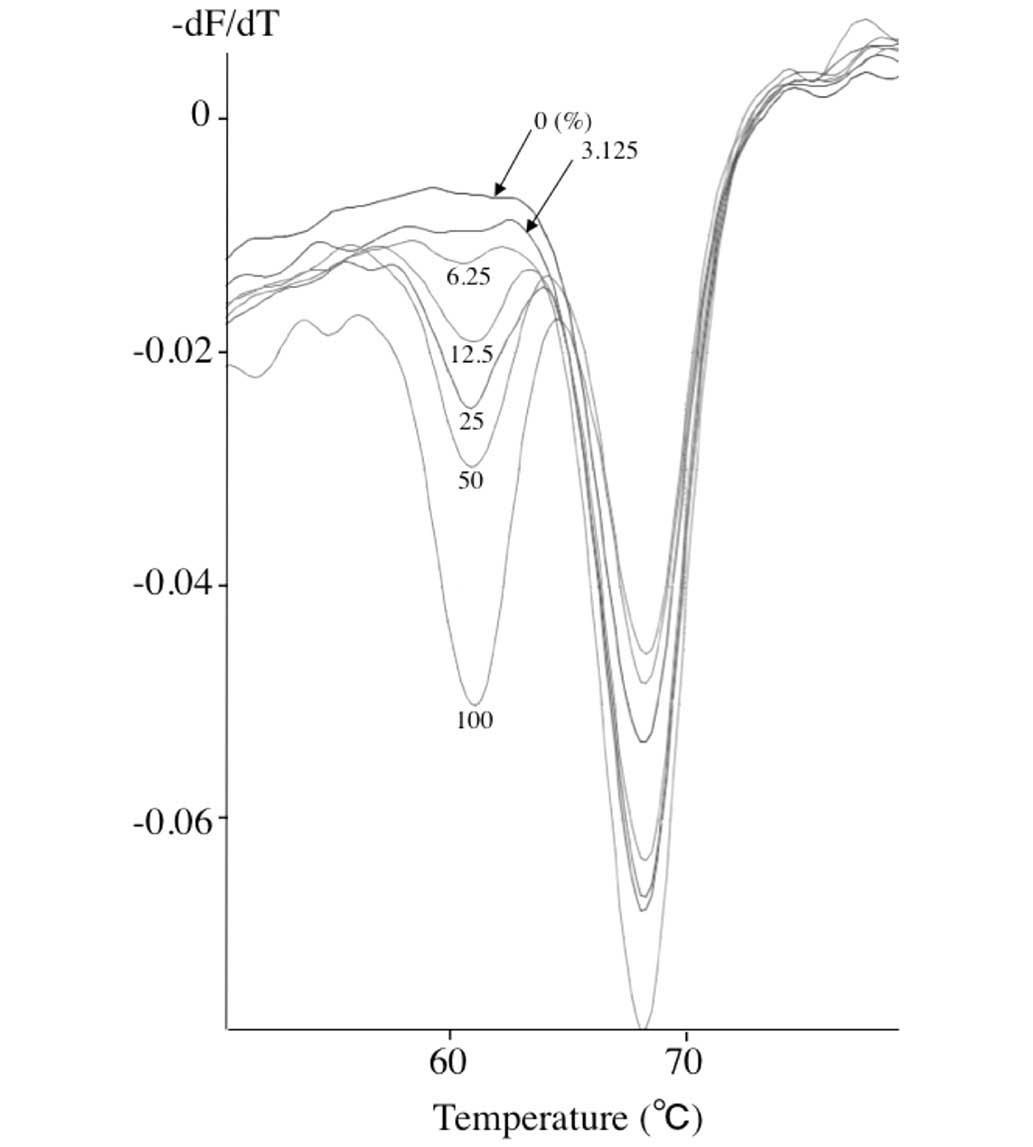
Assay sensitivity. In order to evaluate the
sensitivity of the QP assay, the OCI/AML3 DNA and M-07e DNA were
mixed at various ratios. Fig. 2 shows
the representative results of the lower detection limit obtained
using these mixed samples containing OCI/AML3 DNA and M-07e DNA.
Recognizable curves with the 61°C melt point were obtained from
samples containing as low as 6.25% OCI/AML3 DNA, or 3.125% of
mutant allele.
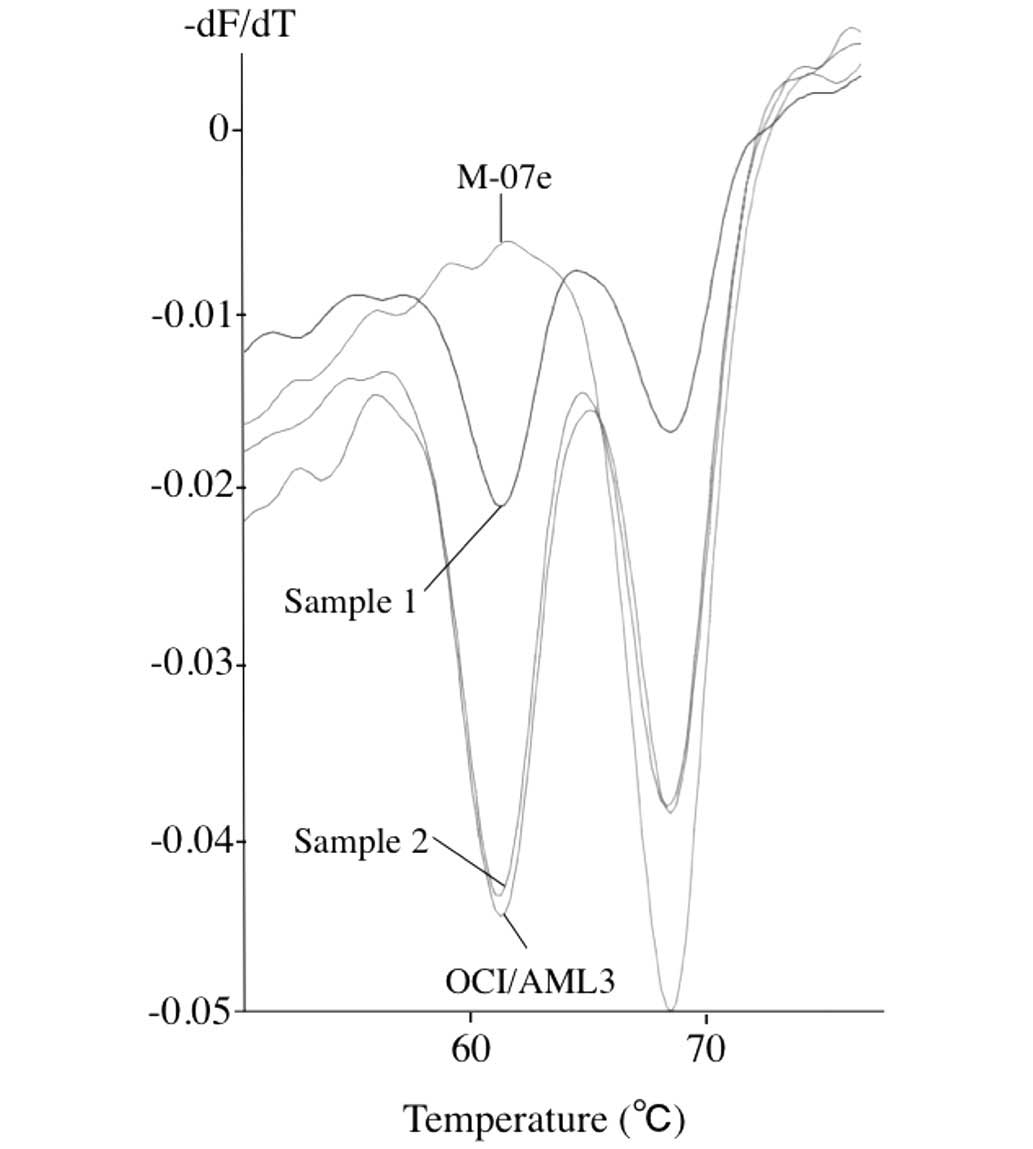
Detection of NPM1 mutations in patients' samples.
Out of the five patient samples examined, three samples presented
the W-shaped curves with lowest points at 61°C and 68°C, indicating
that these samples contained both the mutant alleles and the
wild-type alleles. Fig. 3 shows the
curves of sample 1 and sample 2, both containing wild-type and
mutant alleles. The curve of sample 3 was similar to that of sample
1 (data not shown). Direct sequencing further confirmed that these
three samples have a type A mutant allele (data not shown). The
remaining two samples presented V-shaped curves with lowest points
at 68°C, indicating that these samples contained wild-type allele
only.
Discussion
In the present study, a QP method was established to
detect NPM1 mutations easily with high sensitivity. To the
best of our knowledge, this is the first report to use the QP
method to detect NPM1 mutations. The curve derived from the
mutant allele was easily discriminated from that of the wild-type
allele. Thus far, the authors of the present study performed
screening for NPM1 mutations by gel electrophoresis of PCR
products to detect a band 4-bp longer than the wild-type band;
however, it was not clearly discriminative (data not shown). HRM
analysis of the PCR products to detect the difference of the
melting curves (13) was additionally
performed by the present authors, but again, it was not clearly
discriminative (data not shown). Compared with these two analyses,
the QP method was superior in its specificity.
In terms of efficiency, the QP method took just 2
hours to perform. The sensitivity was quite high as the lower limit
of detection was as low as a 3% concentration of mutant allele. In
clinical settings, NPM1 assays are predominantly used for
diagnosis at presentation, rather than for detection of minimal
residual diseases. The mutant alleles are typically present in at
least 10% of the cells in AML blood or bone marrow samples, even in
heterozygous mutant cases. Therefore, higher sensitivity is not
necessarily needed for diagnosis. Moreover, by comparing the depth
of the two curves as shown in Fig. 2,
the approximate ratio between mutant allele and wild-type allele
can be estimated.
The QP assay also demonstrated its applicability for
clinical use. In the AML patient samples, the mutant allele was
detected in three samples, which were all confirmed to be type A
mutant cases. Non-type A mutant AML samples were not encountered in
this study as the number of available AML samples were few. To
date, at least 61 types of mutant sequences of NPM1 gene
have been reported (2,4–7). The QP 2
sequence used in this assay covers the mutated region of 57 of
these types. If additional samples are available in the future,
further examination as to whether this method can detect these
remaining mutant alleles should be performed. Based on the above
findings, the QP method appears to be an effective tool for
screening NPM1 mutations in AML cases.
Acknowledgements
This work was supported in part by a Grant-in-Aid
for Scientific Research (C) from the Japan Society for the
Promotion of Science (grant no. 18690522).
References
|
1
|
Falini B, Mecucci C, Tiacci E, Alcalay M,
Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E,
Santucci A, et al: Cytoplasmic nucleophosmin in acute myelogenous
leukemia with a normal karyotype. N Engl J Med. 352:254–266. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rau R and Brown P: Nucleophosmin
(NPM1) mutations in adult and childhood acute myeloid
leukaemia: Towards definition of a new leukaemia entity. Hematol
Oncol. 27:171–181. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arber DA, Vardiman JW, Brunning RD, Porwit
A, Le Beau MM, Thiele J, Falni B and Bloomfield CD: Acute myeloid
leukaemia with recurrent genetic abnormalities. WHO classification
of tumours of haematopoietic and lymphoid tissues. Swerdlow SH,
Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J and
Vardiman JW: 2:(4th). (Lyon). IARC Press. 110–123. 2008.
|
|
4
|
Pianta A, Fabbro D, Damiani D, Tiribelli
M, Fanin R, Franzoni A, Romanello M, Tell G, Di Loreto C and
Damante G: Two novel NPM1 mutations in a therapy-responder AML
patient. Hematol Oncol. 28:151–155. 2010.PubMed/NCBI
|
|
5
|
Park SH, Chi HS, Shim H, Jang S and Park
CJ: Two novel NPM1 mutations in an acute myeloid leukemia
patient transformed from primary myelofibrosis. Int J Lab Hematol.
35:e1–e3. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeon Y, Seo SW, Park S, Park S, Kim SY, Ra
EK, Park SS and Seong MW: Identification of two novel NPM1
mutations in patients with acute myeloid leukemia. Ann Lab Med.
33:60–64. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ahmad F, Mandava S and Das BR: Mutations
of NPM1 gene in de novo acute myeloid leukaemia:
Determination of incidence, distribution pattern and identification
of two novel mutations in Indian population. Hematol Oncol.
27:90–97. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Falini B, Nicoletti I, Martelli MF and
Mecucci C: Acute myeloid leukemia carrying cytoplasmic/mutated
nucleophosmin (NPMc+ AML): Biologic and clinical features. Blood.
109:874–885. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Quentmeier H, Martelli MP, Dirks WG, Bolli
N, Liso A, Macleod RA, Nicoletti I, Mannucci R, Pucciarini A,
Bigerna B, et al: Cell line OCI/AML3 bears exon-12 NPM gene
mutation-A and cytoplasmic expression of nucleophosmin. Leukemia.
19:1760–1767. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thiede C, Creutzig E, Illmer T, Schaich M,
Heise V, Ehninger G and Landt O: Rapid and sensitive typing of
NPM1 mutations using LNA-mediated PCR clamping. Leukemia.
20:1897–1899. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oppliger Leibundgut E, Porret NA, Bienz
Muggli M, Baumgartner H, Dahlhaus M and Baerlocher GM: Rapid and
highly specific screening for NPM1 mutations in acute
myeloid leukemia. Ann Hematol. 92:173–177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Scholl S, Mügge LO, Landt O, Loncarevic
IF, Kunert C, Clement JH and Höffken K: Rapid screening and
sensitive detection of NPM1 (nucleophosmin) exon 12 mutations in
acute myeloid leukaemia. Leuk Res. 31:1205–1211. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan AY, Westerman DA, Carney DA, Seymour
JF, Juneja S and Dobrovic A: Detection of NPM1 exon 12 mutations
and FLT3-internal tandem duplications by high resolution melting
analysis in normal karyotype acute myeloid leukemia. J Hematol
Oncol. 1:102008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Laughlin TS, Becker MW, Liesveld JL,
Mulford DA, Abboud CN, Brown P and Rothberg PG: Rapid method for
detection of mutations in the nucleophosmin gene in acute myeloid
leukemia. J Mol Diagn. 10:338–345. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Noguera NI, Ammatuna E, Zangrilli D,
Lavorgna S, Divona M, Buccisano F, Amadori S, Mecucci C, Falini B
and Lo-Coco F: Simultaneous detection of NPM1 and FLT3-ITD
mutations by capillary electrophoresis in acute myeloid leukemia.
Leukemia. 19:1479–1482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ottone T, Ammatuna E, Lavorgna S, Noguera
NI, Buccisano F, Venditti A, Giannì L, Postorino M, Federici G,
Amadori S and Lo-Coco F: An allele-specific RT-PCR assay to detect
type A mutation of the nucleophosmin-1 gene in acute myeloid
leukemia. J Mol Diagn. 10:212–216. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanaka R, Kuroda J, Stevenson W, Ashihara
E, Ishikawa T, Taki T, Kobayashi Y, Kamitsuji Y, Kawata E, Takeuchi
M, et al: Fully automated and super-rapid system for the detection
of JAK2V617F mutation. Leuk Res. 32:1462–1467. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ono A, Okuhashi Y, Takahashi Y, Itoh M,
Nara N and Tohda S: Advantages of the quenching probe method over
other PCR-based methods for detection of the JAK2 V617F
mutation. Oncol Lett. 4:205–208. 2012.PubMed/NCBI
|
|
19
|
Kurata S, Kanagawa T, Yamada K, Torimura
M, Yokomaku T, Kamagata Y and Kurane R: Fluorescent quenching-based
quantitative detection of specific DNA/RNA using a BODIPY((R))
FL-labeled probe or primer. Nucleic Acids Res. 29:E342001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rothberg PG, Ramirez-Montealegre D,
Frazier SD and Pearce DA: Homogeneous polymerase chain reaction
nucleobase quenching assay to detect the 1-kbp deletion in CLN3
that causes Batten disease. J Mol Diagn. 6:260–263. 2004.
View Article : Google Scholar : PubMed/NCBI
|