Introduction
Colorectal cancer (CRC) is a common gastrointestinal
malignancy, and worldwide, it is ranked the third most frequent
type of cancer in men and the second most frequent type in women.
In North America, New Zealand, Western Europe, Japan and other
economically developed areas, CRC ranks first or second in
frequency among all cancers in men. The clinical presentation of
CRC and the associated mortality has followed an increasing trend
(1). Hypoxia is a common pathological
feature of solid tumors, such as CRC, and is caused by low levels
of tumor cell differentiation and rapid tumor growth with
insufficient growth of blood vessels, resulting in an insufficient
blood supply to the tumor (2).
Hypoxia induces the high expression of
hypoxia-inducible factor-1α (HIF-1α). A previous study identified
that ~55% of CRC patients exhibit elevated expression levels of
HIF-1α (3). Activated HIF-1α binds to
the hypoxia response element and forms the response element binding
protein with other transcription factors, to regulate the
expression of ~60 target genes. These factors include vascular
endothelial growth factor (VEGF) and basic fibroblast growth
factor, which are now recognized as the most important
angiogenesis-promoting factors (4).
HIF-1α expression is important in the development of CRC, and
transient HIF-1α expression in colon cancer cells can rapidly
increase VEGF expression (5).
Furthermore, overexpression of HIF-1α in CRC cells injected
subcutaneously into nude mice can promote cell proliferation and
angiogenesis (6). As hypoxia can
induce the epithelial-mesenchymal transition of CRC cells, thus
promoting metastasis (7),
hypoxia-induced gene expression profiles may be used as independent
prognostic markers for predicting the outcome of stage II and III
colon cancers (8).
Hypoxia is important in promoting CRC occurrence,
angiogenesis and metastasis. It is important to identify and verify
the key gene that is responsible for regulation of tumor
angiogenesis and metastasis following activation of HIF-1α. The
present study aimed to investigate the practical significance of
HIF-1α targeting for the treatment and prevention of CRC
angiogenesis, invasion and metastasis. A lentiviral vector
containing green fluorescent protein (GFP) and HIF-1α was
constructed and used to infect CRC cell lines in vitro, for
coexpression of these proteins in stably-transfected cell lines.
Establishing a visual xenograft nude mouse model may lay the
foundation for studying the biological functions of HIF-1α in CRC
in vivo.
Materials and methods
Materials
The lentiviral vectors pLV-TRC-EGFP, pΔ8.91 (virus
helper plasmid) and pMD2.G (helper plasmid, encoding VSV-G), and
293T cells were purchased from Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd. (Beijing, China). The human CRC SW480,
SW620, LoVo and HCT116 cell lines were purchased from the cell bank
of the Shanghai Institute of Biochemistry and Cell Biology
(Shanghai, China). Tissue culture flasks and 96-well plates were
purchased from Greiner Bio-One GmbH (Frickenhausen, Germany).
Minimum essential medium (MEM) and Dulbecco's modified Eagle's
medium (DMEM) were purchased from Life Technologies (Grand Island,
NY, USA), and X-tremeGENE transfection reagent was purchased from
Roche Diagnostics (Basel, Switzerland). The plasmid miniprep kit
and molecular cloning enzymes were purchased from Sangon Biotech
Co., Ltd. (Shanghai, China). NheI was obtained from New
England BioLabs, Inc. (Ipswich, MA, USA) and agarose gel was
purchased from Biowest (Nuaillé, France). High-fidelity Taq
polymerase was purchased from Toyobo Co., Ltd., (Osaka, Japan), LB
medium was obtained from Caisson Laboratories (Logan, UT, USA) and
puromycin, polybrene and ampicillin were purchased from
Sigma-Aldrich (St. Louis, MO, USA). Antibodies used for western
blotting were purchased from Santa Cruz Biotechnology, Inc.,
(Dallas, TX, USA) and the nude mice was purchased from Changzhou
Card Vince Laboratory Animal Co., Ltd. (Luoyang, China).
Construction of overexpressing
lentiviral vector
i) Primers targeting the HIF-1α open reading frame
(ORF) of the pBabe vector (obtained from Professor Ji Hongbin,
Laboratory of the Shanghai Institute of Biochemistry and Cell
Biology, Shanghai, China) were designed with an NheI
restriction site at the 5′ end. Primers were designed using Vector
NTI® software version 10.0 (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), which covered the open reading frame of HIF1α.
High-fidelity Taq polymerase was used for polymerase chain
reactions (PCR) to amplify the ORF DNA fragment, and the DNA
template was removed using DpnI. Following digestion with
NheI, the purified PCR product was ligated to the
pLV-TRC-EGFP plasmid. ii) DH5α competent cells: 2 µl of the
ligation products were transformed into 25 µl Escherichia
coli cells (~5×107 cell/ml) with DH5α that had been
plated on a petri dish containing 100 g/ml ampicillin in lysogeny
broth (LB) medium and incubated at 37°C overnight. Three monoclonal
colonies were picked from each dish and inoculated into 3 ml LB
medium with ampicillin, and then incubated overnight at 37°C on a
shaker. iii) Identification of the recombinant plasmid: The plasmid
was extracted using a miniprep kit (Qiagen GmbH, Hilden, Germany),
and identified by digestion with NheI. The plasmid was
identified using 1% agarose gel (Biowest, Nuaillé, France)
electrophoresis with electrophoresis apparatus (HE120
Multifunctional Horizontal Gel Electrophoresis Cell; Tanon Science
& Technology Co., Ltd., Shanghai, China), and the present study
refers to this plasmid as pLV-HIF1α-EGFP. iv) Sequencing of
expression vectors: Digestion using the appropriate restriction
enzymes confirmed the correct fragments were isolated and amplified
from bacterial cultures, and these were sent for sequencing
(Biosune Biotechnology Co., Ltd., Shanghai, China). v) Large-scale
plasmid extraction: The sequenced pLV-HIF1α-EGFP plasmid was
amplified in LB broth using a Plasmid kit (Qiagen, Inc., Valencia,
CA, USA) for large-scale plasmid extraction, and the isolated
plasmids were subjected to quality control. Isolated plasmids were
examined using a NanoDrop 2000™ spectrophotometer (Thermo Fisher
Scientific, Inc.), and an optical density value of 1.8 (at 260/280
nm) was treated as the quality control.
Lentiviral particles packaging and
titer testing
The 293T cells were plated onto tissue culture the
day prior to transfection. On the day of transfection, cell
confluency did not exceed 80%. The X-tremeGENE reagent was used to
transfect the expression plasmid containing pLV-HIF1α-EGFP and the
two packaging plasmids pΔ8.91 and pMD2.G into the 293T cells,
according to a mass ratio of 10:10:1. Following 12 h of incubation
at 37°C, the complete medium was replaced, and after 24 h of
incubation, expression of GFP was observed in the 293T cells, using
a fluorescence microscope (Axio Imager 2; Zeiss, Oberkochen,
Germany). Cell culture supernatants were collected during 48–72 h,
and virus particles were passed through a 0.45-µm filter and stored
at 4°C. The purified virus particles were titred as follows: 293T
cells in the logarithmic growth phase were plated onto 96-well
plates (8,000 cells/well) and cultured at 37°C in DMEM containing
2% fetal bovine serum (FBS; Biological Industries, Beit-Haemek,
Israel) until 30–50% confluency was reached, after 16 h. The
purified virus suspension was diluted 1:10 with DMEM containing 2%
FBS, and used to replace the medium in the 96-well plates. Each
virus dilution was used to infect two wells. Following incubation
overnight at 37°C, the virus-containing medium was discarded and
replaced with DMEM containing 10% FBS. At day 5, the number of
fluorescent cells in each virus dilution was observed under a
fluorescence microscope. Transducing units (TU) were calculated as
follows: Titer = cells with fluorescence × dilution ratio, and the
final lentivirus titer was 5×107 TU/ml.
Lentiviral infection
CRC cells in the logarithmic growth phase were
seeded into 6-well plates. Following 12 h of culture, the
supernatant was discarded and 200 µl/well of virus suspension was
added to medium containing polybrene (concentration is 4 µg/m1). At
8 h post-viral infection, the medium was changed to DMEM with 2%
FBS. Following 48 h of cell culture, puromycin (0.5 µg/ml) was
added to the medium. The culture medium was replaced every 2 days
to remove dead cells. After screening with 0.5 µg/ml puromycin
three times to determine which cells had undergone successful
transfection, DMEM containing 10% FBS was added for 24 h and the
cells were then collected in culture flasks for subsequent
experiments.
Western blot analysis
The cells were rinsed twice with cold
phosphate-buffered saline (PBS) and then lysed on ice in
radioimmunoprecipitation assay lysis buffer. Following this, the
cells were collected and the proteins were separated on a 10%
acrylamide gel using sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and a Bio-Rad mini system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Separated proteins were then transferred
to a polyvinylidene difluoride membrane (EMD Millipore, Billerica,
MA, USA). Non-specific antibody binding sites were blocked for 1 h
with 5% skimmed milk (Nestlé, Vevey, Switzerland). Anti-HIF-1α
mouse monoclonal anti-human (cat. no. ab82832; dilution, 1:1,000;
Abcam, Cambridge, MA, USA), anti-VEGF rabbit polyclonal anti-human
(cat. no. ab46154; dilution, 1:1,000; Abcam) and anti-pyruvate
kinase isozyme M1 (PKM1) rabbit polyclonal anti-human (cat. no.
ab156849; dilution, 1:1,000; Abcam) primary antibodies were
incubated with the membrane at 4°C overnight. Subsequent to being
washed with Tris-buffered saline plus Tween 20 (TBST), the
horseradish peroxidase-conjugated rabbit anti-mouse (cat. no. 7076;
dilution, 1:3,000; Cell Signaling Technology, Inc., Danvers, MA,
USA) and goat anti-rabbit (cat. no. 7074; dilution, 1:3,000; Cell
Signaling Technology) IgG secondary antibodies were added at room
temperature for 1 h. The membrane was then washed three times with
TBST. Electrochemiluminescence reagent was then applied to the
membrane (Life Technologies), and film was developed using a
darkroom.
Cell migration assay
Matrigel (stored at −80°C) was allowed to melt at
room temperature, and was then diluted 1:4 with MEM. A total of 50
µl of the solution was added to each Transwell chamber (Corning
Inc., Corning, NY, USA) and allowed to dry at 4°C. The excess
solution was absorbed and 50 µl of medium was added to each chamber
and incubated at 37°C for 30 min. The cells were suspended at a
density of 5×105 cells/ml in MEM. Cell suspension (100
µl) was placed in each upper chamber, and 600 µl MEM containing 10%
FBS was placed into the lower compartment. Following incubation for
3–6 h, the cell culture medium of the upper and lower chambers was
discarded and the membrane was removed and dried. Crystal violet
solution (600 µl; diluted 1:4 in 2% ethanol) was applied to the
membranes for 30 min. The membranes were washed twice with PBS, and
the upper surface of each membrane was wiped with a cotton swab to
remove non-migrated cells. The cells that had migrated through the
membrane were visualized using a microscope (CX21BIM-SET5; Olympus
Corp., Tokyo, Japan).
Establishment of an abdominal tumor
nude mouse model
Logarithmic growth phase CRC cells were digested
with 0.25% trypsin and washed once with fresh medium. The cell
pellet was collected by centrifugation at 1,000 × g for 3 min and
the supernatant was discarded. The cells were washed twice with
sterile saline and counted. The concentration of live cells was
adjusted to 1×107/ml. Prior to inoculation, the skin of
the nude mice was disinfected with alcohol. The cell suspension
(200 µl) was injected intraperitoneally into mice with a 1-ml
syringe, to confer a cell mass of 2×106 cells/mouse.
After 14 days, the mice were dissected to remove any nodules and to
confirm the expression of GFP under a fluorescence microscope.
Statistical analysis
All data are expressed as the mean ± standard
deviation. The paired Student's t-test and χ2 test were
used to determine statistical significance. Statistical analysis
was performed with GraphPad Prism software version 5 (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
HIF-1α vector construction,
identification and overexpression
The pLV lentiviral vector was used to stabilize
expression of GFP and HIF-1α by molecular cloning. Molecular
analysis of the plasmid identified two (8.8-kbp and 2.5-kbp) bands,
which were cut out following incubation with the NheI
restriction enzyme. The correctly identified plasmid was sent to
Biosune Biotechnology Co., Ltd. for nucleotide sequencing using
universal primers, and this confirmed that the correct nucleotide
sequence was present. Thus, the constructed plasmid could be used
for the transfection of the 293T cells and the expression of the
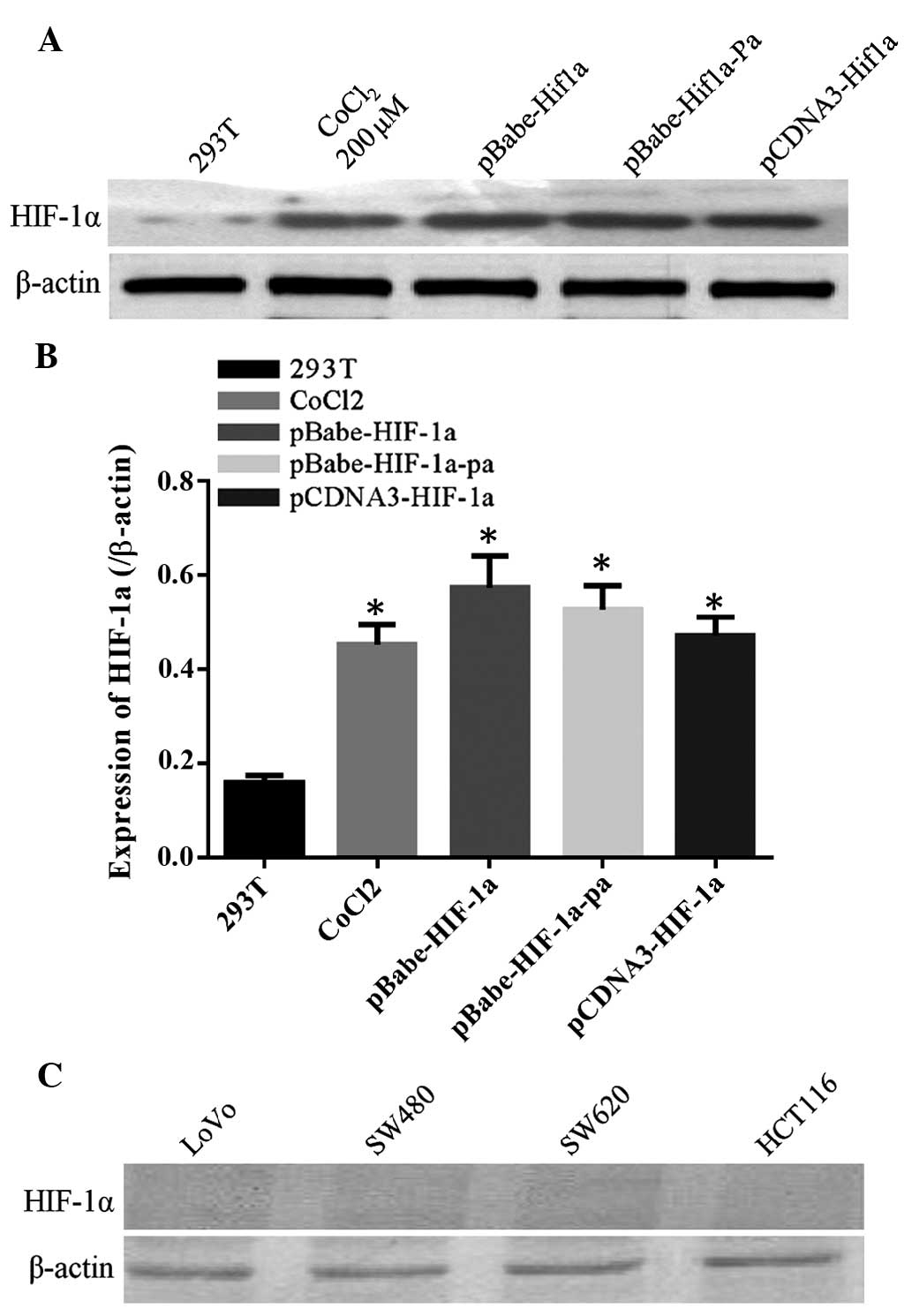
coding sequence of HIF-1α isoforms 1 and 3 (Fig. 1).
HIF-1α expression levels in CRC
HIF-1α expression was not detected under normal
conditions in four different CRC cell lines, as shown in Fig. 2. Previous studies have indicated that
CoCl2 can simulate cell hypoxia and induce the
expression of HIF-1α (9). Fig. 3 demonstrates that the SW620 and HCT116
cell lines had elevated levels of HIF-1α following treatment with
CoCl2 for 24 h, while HIF-1α expression in the LoVo
cells was only slightly upregulated. HIF-1α expression in the SW480
cells was not induced, because that SW480 was the low metastatic
potential CRC cell lines, SW480 as a targeted infection cell lines
for lentiviral infection after packaging could be better observed
the effect of HIF-1α overexpression on tumor biology.
HIF-1α overexpression plasmid
transfection and lentivirus packaging
The constructed pLV-HIF1α-GFP plasmid and the
corresponding helper plasmid were used to transfect 293T cells
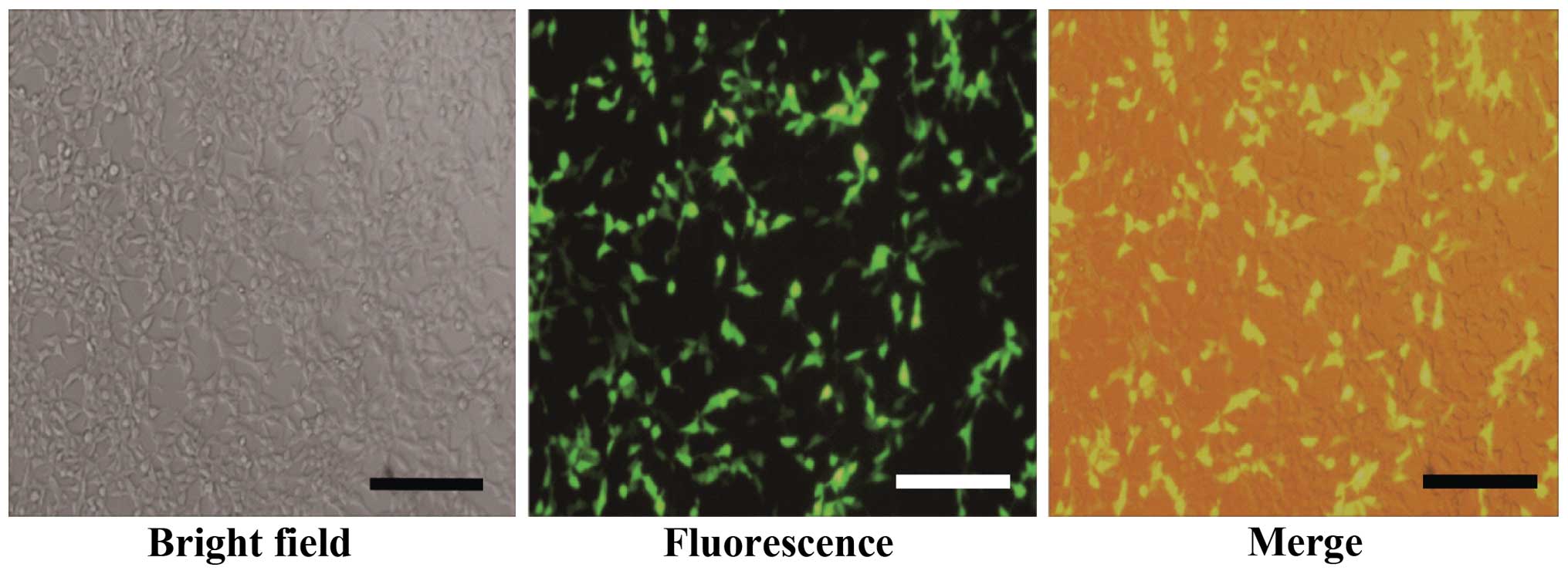
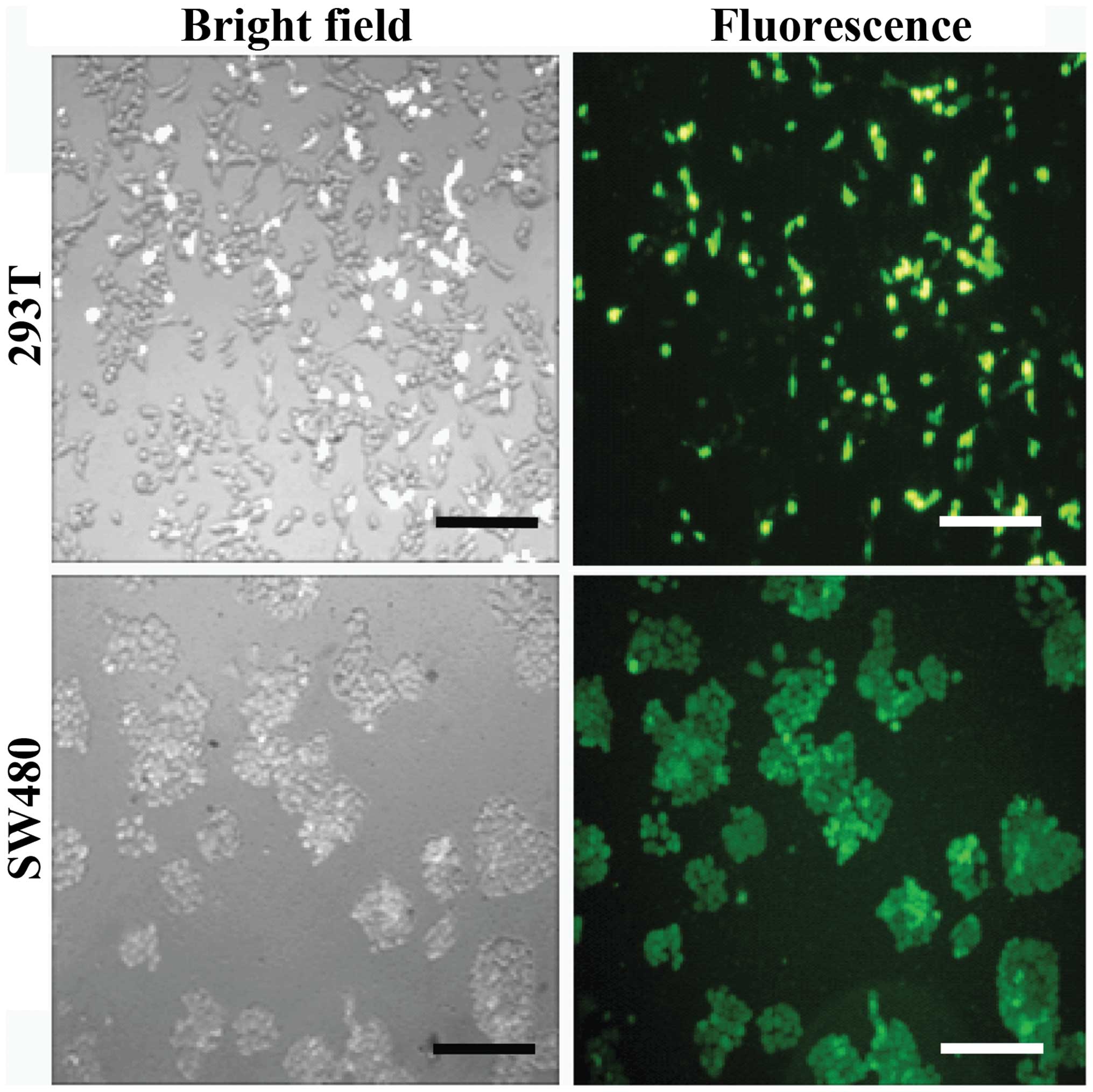
(Fig. 4). The transfection efficiency
was >80%. The culture supernatant virus was collected for titer
determination after transfected 48 h and the virus titer was
5–7×106 transduction units (TU)/ml. After the SW480
cells were infected with lentivirus for 72 h, the expression of GFP
was observed by fluorescence microscopy and cell morphology was
analyzed, confirming effective lentivirus infection of the SW480
cells and expression of the target protein (Fig. 5). Next, puromycin was added, and after
3 weeks of screening, cell lines stably expressing HIF-1α were
obtained. Western blot analysis confirmed the screened stable
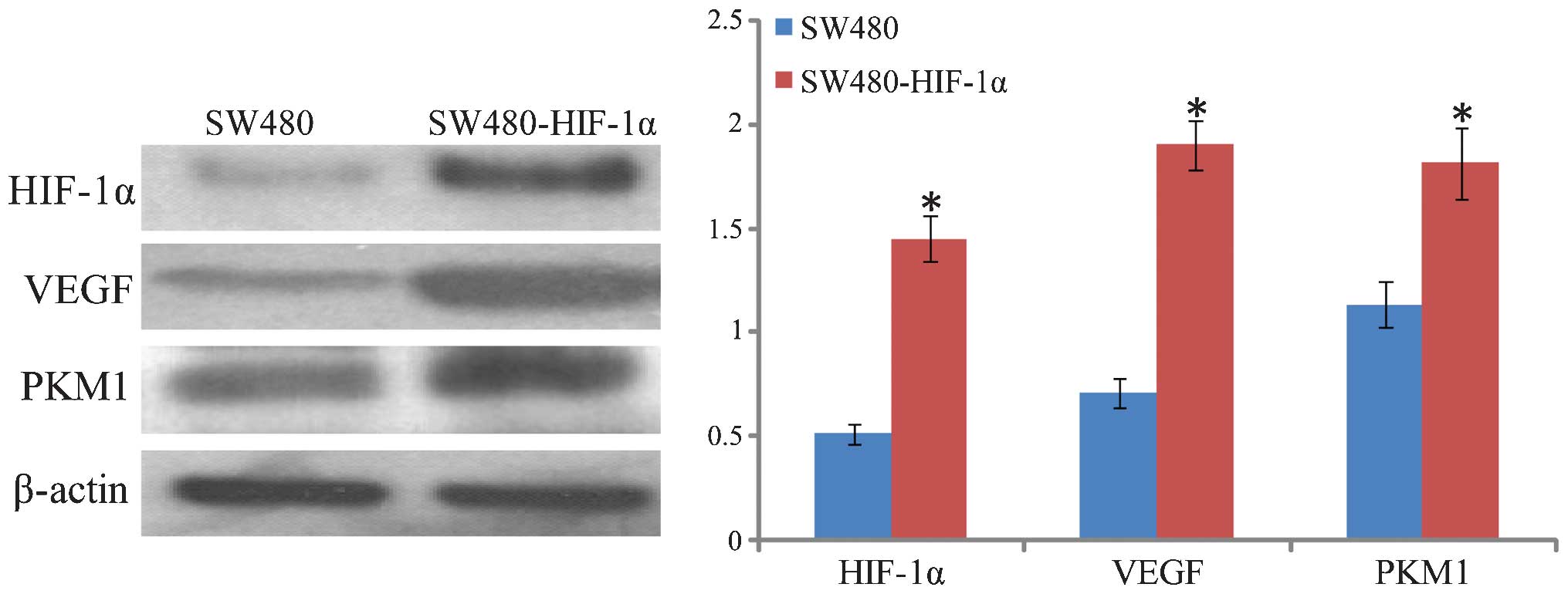
SW480-HIF-1α was successfully constructed (Fig. 6).
Stable HIF-1α expression in SW480
cells following puromycin screening
Western blot analysis confirmed that HIF-1α was
highly expressed in the SW480 cells following puromycin screening
(Fig. 6). In addition, the HIF-1α
downstream target genes, VEGF and PKM1, exhibited a certain degree
of increased expression. VEGF exhibited the most significant
upregulation, followed by PKM1, suggesting that the overexpression
of HIF-1α may have an extensive role in cells, inducing
angiogenesis growth factors and promoting glycolysis.
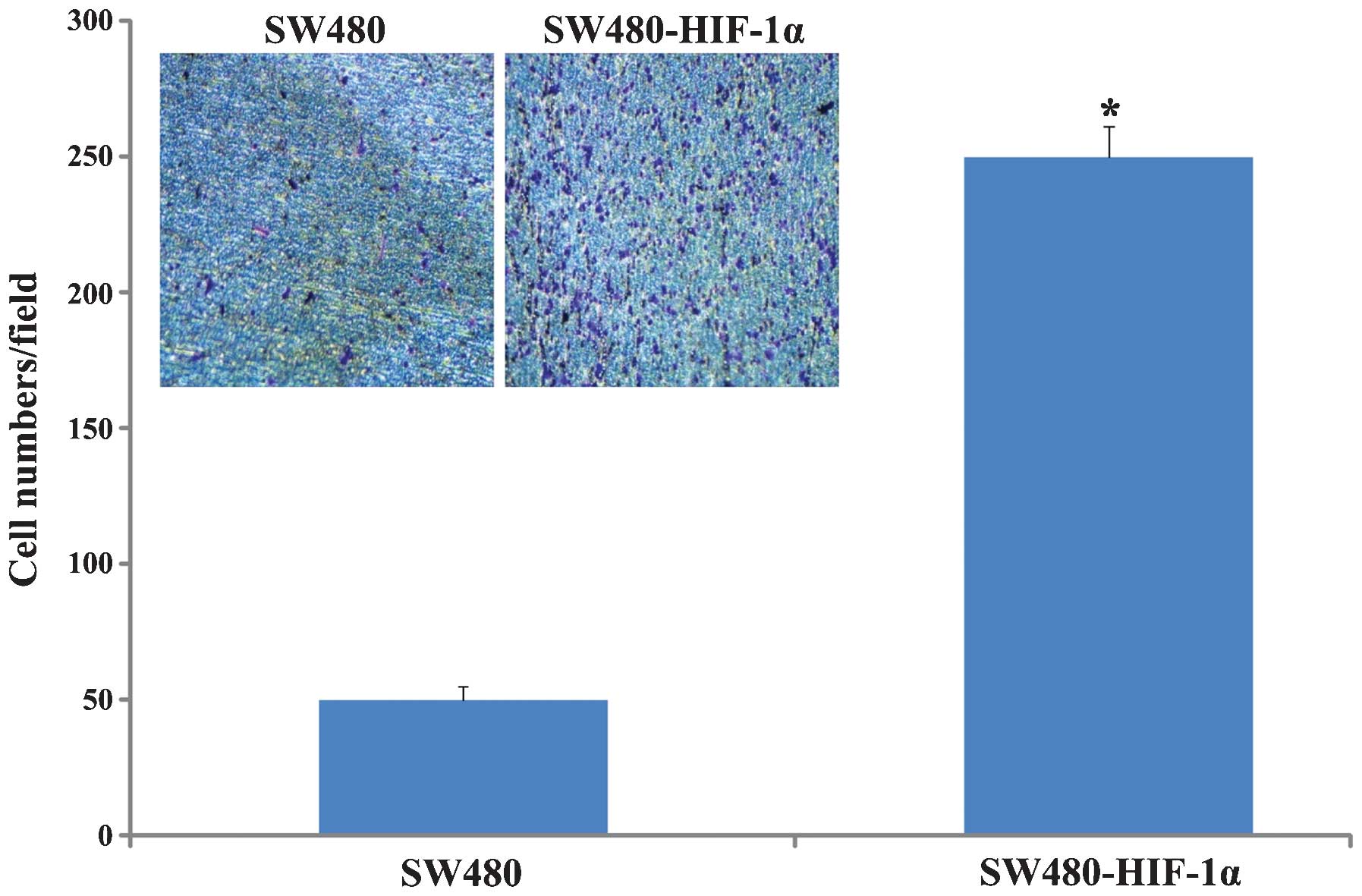
Effects on cell migration after HIF-1α
stable expression
Transwell chamber mobility change was comparable
prior to and following HIF-1α overexpression; the results showed
that prior to SW480-HIF-1α stable overexpression, mobility had
improved significantly (P<0.0001; Fig.
7), increased from 50±5 cells/field to 250±11 cells/field (x200
magnification). These data suggested that HIF-1α overexpression
significantly increased the invasion and migration of cells.
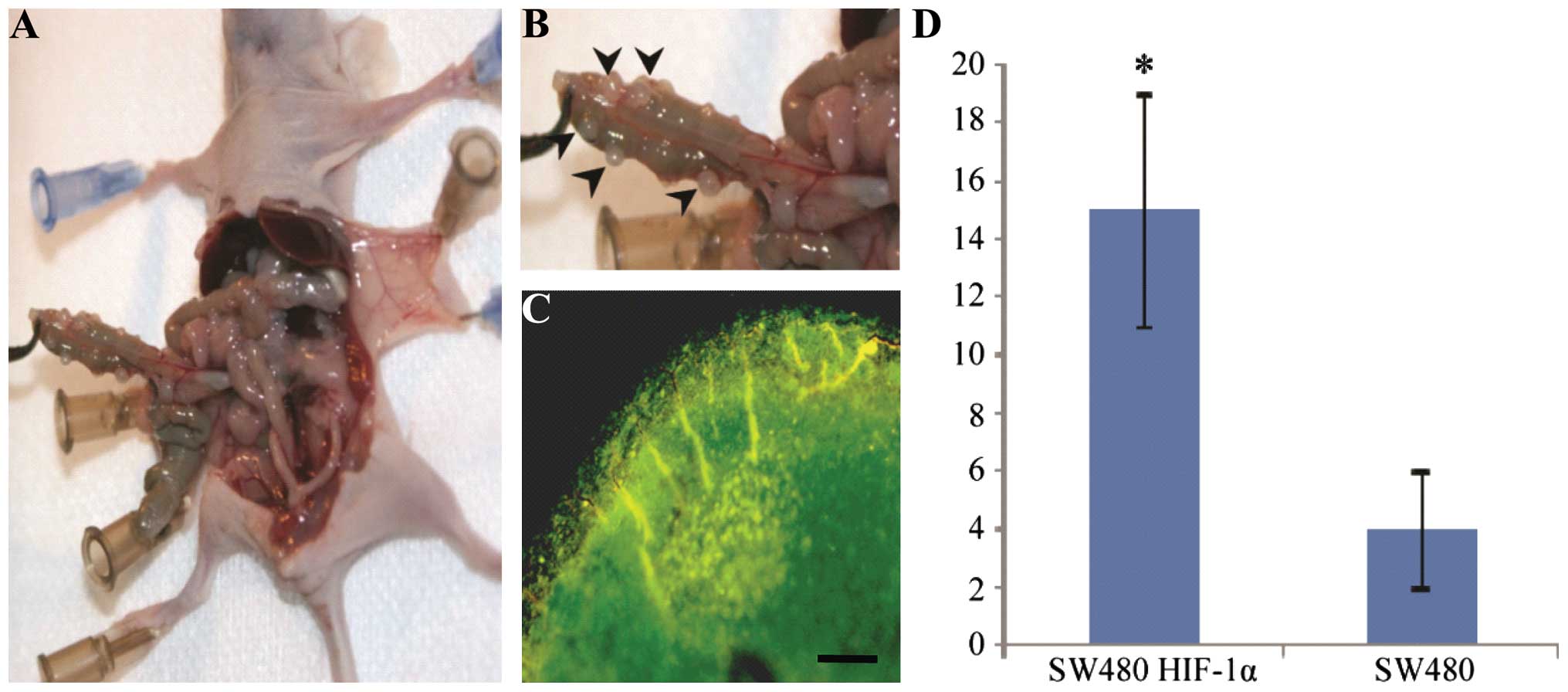
Construction of GFP/HIF-1α dual
expression xenografts in nude mice
CRC cells expressing GFP and HIF-1α were injected
into the abdomen of nude mice. After 14 days, nodules were
dissected from the nude mice and counted (Fig. 8), Mice with SW480-HIF-1α exhibited
nude multiple nodules in the abdominal cavity (Fig. 8A and B), with 15±4 nodules/mouse
compared with 4±1 nodules/mouse in the control group; this
difference was statistically significant (P<0.05). Subsequent to
removal of the nodules, they were observed under a fluorescence
microscope; nodules with green fluorescence expressed high levels
of GFP and HIF-1α, present in the SW480 colon cancer cells
(Fig. 8C).
Discussion
Gene expression in lentivirus-infected target cells
is stable and persistent (10). Since
the first use of the human immunodeficiency virus as a gene
transfer vector, the ability has existed to integrate a pseudovirus
into the host genome. The expressed gene is not affected by cell
division or the environment (11),
therefore, the technique can significantly increase the expression
levels of unstable proteins in cells (10). This technology constitutes a more
efficient class of expression vectors, which not only aid basic
cancer research, but can also be used for gene therapy in the
clinic (12). HIF-1α is an important
factor in tumorigenesis and development, and there is extensive
systematic research into its role in CRC. HIF-1α is only expressed
under hypoxic conditions, therefore, experimental study of its
functions requires the use of hypoxic conditions or
CoCl2, which simulates hypoxia in vitro. To
generate hypoxic conditions for cell culture, researchers must use
a humidified incubator with varying proportions of O2
and CO2 or a low oxygen chamber (O2 may be
downregulated to 3% using a multi-gas inhibitor) (13). These requirements add to the
experimental cost and amount of apparatus required, as well as
inducing a cellular stress response. In addition to the expression
of HIF-1α, other proteins exist that are regulated by a stress
response feedback. The use of high concentrations of
CoCl2 may change the osmotic pressure in cells, and can
exert other effects, such as inhibiting tumor cell proliferation
(14). Thus, the traditional methods
for inducing the expression of HIF-1α are associated with a number
of complicating factors that have made the elucidation of gene
function more difficult (15); this
may be the reason for the low availability of drugs currently
targeting HIF-1α (16).
HIF-1α protein is abundantly expressed the majority
of solid tumors, including colon, breast, pancrease, kidney,
prostate, ovary, brain and bladder cancer (6,17,18). In turn, inhibiting HIF-1α may directly
inhibit tumor formation (19), which
is not conducive for studying the in vivo function of
HIF-1α. Establishing GFP and HIF-1α stable co-expression in CRC
cell lines using lentiviral technology may solve the aforementioned
problems. Furthermore, as HIF-1α is hardly expressed in the
normoxic state in tumor studies, it is vital to establish cell and
animal models with with expression of HIF-1α under normal
conditions. The establishment of models in vivo and in
vitro may improve our understanding of the role of HIF-1α in
tumors and additionally help with the identification of
HIF-1α-targeted drugs. In addition, via the establishment of cells
that demonstrate co-expression of HIF-1α and GFP, the role of
HIF-1α in regulation of proliferation and invasion of tumor cells
may be observed.
The present study constructed a lentivirus with
stable HIF-1α expression, and screened the stable SW480-HIF1α cell
line. Transwell migration assays demonstrated that following
overexpression of HIF-1α, cell migration and proliferation were
enhanced. This is in agreement with the results of previous studies
that have investigated HIF-1α in cancer cells (20). In the present study, following
intraperitoneal injection into nude mice, the cells formed large
tumor nodules, and these nodules were identified as resulting from
SW480-HIF-1α cells by using fluorescence microscopy. This method
provides a novel in vivo model of HIF-1α, and in future
studies, the tracking of GFP fluorescence may indicate the relevant
tumor localization in vivo.
The present study method has certain limitations,
for example, the process of vector construction and validation was
time-consuming. The main steps included obtaining cDNA,
stably-expressing HIF-1α plasmid construction, lentivirus packaging
and infection, stable cell line screening and tumorigenicity
analysis. This multi-stage process is not conducive for large-scale
laboratory applications. It is possible that diverse cDNA sequences
could be obtained through collaboration and commercialization of
laboratories in the future, thus reducing the amount of time
required for the cloning procedure. Furthermore, during the
insertion of exogenous fragments, due to the limitations of the
lentiviral vector itself, the restriction sites of the enzyme
multiple cloning site were relatively small, and this made it
difficult to choose target sites for the primers. To facilitate
clone construction, more unique rare restriction site for enzymes
could be arranged, including AscI, SwaI and
PmeI, and other cleavage sites. Further improvements to the
efficiency of transfection would significantly shorten the
screening time of stable cells by virus infection, and thus reduce
the overall testing process. The present study did not use the
conventional calcium phosphate transfection method, mainly as
calcium phosphate transfection reagent is generally prepared by
onsite laboratory personnel themselves, with a lack of appropriate
quality control procedures, and also due to the different
processing techniques used to prepare phosphate-plasmid complexes
causing a relatively large difference in the size of the formed
particles, thus an unstable transfection efficiency and unreliable
results.
The small animal in vivo imaging system was a
novel animal imaging system introduced in recent years, which could
be used for the location of fluorescent labeled compounds,
peptides, cells and nucleic acids in rats or mice. However, as
small animal in vivo imaging exhibited a poor resolution for
GFP, the green fluorescence produced could not be effectively
detected through the skin of nude mice and there were
autofluorescence interference problems at the skin surface.
Therefore, in the present study, the nature of the nodules was
determined after the mice were dissected, as good imaging data
could not be obtained using the small animal imaging system. The
techniques used in the present study are outdated compared with
those used in a previous study, which reported the use of GFP to
obtain in vivo imaging (21).
Subsequent studies should focus on substituting luciferase for GFP
in the plasmids. Following injection of the luciferase substrate
into mouse models, the cold light could be motivated more easily to
penetrate animal skin, and is more sensitive compared with the GFP
method used in the present study (22). Future studies could be focused on
investigating the downstream proteins in the HIF-1α cascades and
clarifying the role of these proteins in tumor proliferation, cell
cycle processes and tumor invasion.
In conclusion, the lentiviral technology employed in
the present study achieved the stable expression of HIF-1α, an
important protein in the development of CRC, and a GFP tag, which
allows for observation of the protein in vivo and in
vitro. Lentiviral technology may provide reliable expression of
proteins for the study of chronic and acute hypoxia-related
diseases, such as stroke (23), high
altitude pulmonary edema and obstructive sleep apnea syndrome, as
well as in vivo models of disease.
Acknowledgements
The present study was funded and supported by the
National Natural Science Foundation of China (grant no. 51003078)
and Shanghai Science and Technology Special Funding for Laboratory
Animals (grant no. 12140902302).
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nishimoto A, Kugimiya N, Hosoyama T, Enoki
T, Li TS and Hamano K: HIF-1α activation under glucose deprivation
plays a central role in the acquisition of anti-apoptosis in human
colon cancer cells. Int J Oncol. 44:2077–2084. 2014.PubMed/NCBI
|
|
3
|
Cao D, Hou M, Guan YS, Jiang M, Yang Y and
Gou HF: Expression of HIF-1α and VEGF in colorectal cancer:
Association with clinical outcomes and prognostic implications. BMC
Cancer. 9:4322009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SE, Shim KN, Jung SA, Yoo K and Lee
JH: The clinicopathological significance of tissue levels of
hypoxia-inducible factor-1α and vascular endothelial growth factor
in gastric cancer. Gut Liver. 3:88–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lucarini G, Zizzi A, Belvederesi L,
Kyriakidou K, Mazzucchelli R and Biagini G: Increased VEGF165
expression in HCT116 colon cancer cells after transient
transfection with a GFP vector encoding HIF-1 gene. J Exp Clin
Cancer Res. 26:515–519. 2007.PubMed/NCBI
|
|
6
|
Zhong H, De Marzo AM, Laughner E, Lim M,
Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL and Simons
JW: Overexpression of hypoxia-inducible factor 1α in common human
cancers and their metastases. Cancer Res. 59:5830–5835.
1999.PubMed/NCBI
|
|
7
|
Hongo K, Tsuno NH, Kawai K, Sasaki K,
Kaneko M, Hiyoshi M, Murono K, Tada N, Nirei T, Sunami E, et al:
Hypoxia enhances colon cancer migration and invasion through
promotion of epithelial-mesenchymal transition. J Surg Res.
182:75–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dekervel J, Hompes D, van Malenstein H,
Popovic D, Sagaert X, De Moor B, Van Cutsem E, D'Hoore A, Verslype
C and van Pelt J: Hypoxia-driven gene expression is an independent
prognostic factor in stage II and III colon cancer patients. Clin
Cancer Res. 20:2159–2168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ciafrè SA, Niola F, Giorda E, Farace MG
and Caporossi D: CoCl(2)-simulated hypoxia in skeletal muscle cell
lines: Role of free radicals in gene up-regulation and induction of
apoptosis. Free Radic Res. 41:391–401. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cockrell AS and Kafri T: Gene delivery by
lentivirus vectors. Mol Biotechnol. 36:184–204. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ciuffi A: Mechanisms governing lentivirus
integration site selection. Curr Gene Ther. 8:419–429. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McGarrity GJ, Hoyah G, Winemiller A, Andre
K, Stein D, Blick G, Greenberg RN, Kinder C, Zolopa A,
Binder-Scholl G, et al: Patient monitoring and follow-up in
lentiviral clinical trials. J Gene Med. 15:78–82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kubis HP, Hanke N, Scheibe RJ and Gros G:
Accumulation and nuclear import of HIF1 alpha during high and low
oxygen concentration in skeletal muscle cells in primary culture.
Biochim Biophys Acta. 1745:187–195. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang B, Guo W, Yu L, Wang F, Xu Y, Liu Y
and Huang C: Cobalt chloride inhibits tumor formation in
osteosarcoma cells through upregulation of HIF-1α. Oncol Lett.
5:911–916. 2013.PubMed/NCBI
|
|
15
|
Wang V, Davis DA, Haque M, Huang LE and
Yarchoan R: Differential gene up-regulation by hypoxia-inducible
factor-1 alpha and hypoxia-inducible factor-2 alpha in HEK293T
cells. Cancer Res. 65:3299–3306. 2005.PubMed/NCBI
|
|
16
|
Onnis B, Rapisarda A and Melillo G:
Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med.
13:2780–2786. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Talks KL, Turley H, Gatter KC, Maxwell PH,
Pugh CW, Ratcliffe PJ and Harris AL: The expression and
distribution of the hypoxia-inducible factors HIF-1alpha and
HIF-2alpha in normal human tissues, cancers, and tumor-associated
macrophages. Am J Pathol. 157:411–421. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Buffa FM, West C, Byrne K, Moore JV and
Nahum AE: Radiation response and cure rate of human colon
adenocarcinoma spheroids of different size: The significance of
hypoxia on tumor control modelling. Int J Radiat Oncol Biol Phys.
49:1109–1118. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ryan HE, Lo J and Johnson RS: HIF-1 alpha
is required for solid tumor formation and embryonic
vascularization. EMBO J. 17:3005–3015. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tsai YP and Wu KJ: Hypoxia-regulated
target genes implicated in tumor metastasis. J Biomed Sci.
19:1022012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li ZH, Liao W, Cui XL, Zhao Q, Liu M, Chen
YH, Liu TS, Liu NL, Wang F, Yi Y, et al: Intravenous
transplantation of allogeneic bone marrow mesenchymal stem cells
and its directional migration to the necrotic femoral head. Int J
Med Sci. 8:74–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choy G, O'Connor S, Diehn FE, Costouros N,
Alexander HR, Choyke P and Libutti SK: Comparison of noninvasive
fluorescent and bioluminescent small animal optical imaging.
Biotechniques. 35:1022–1026, 1028-1030. 2003.PubMed/NCBI
|
|
23
|
Ralph GS, Parham S, Lee SR, Beard GL,
Craigon MH, Ward N, White JR, Barber RD, Rayner W, Kingsman SM, et
al: Identification of potential stroke targets by lentiviral vector
mediated overexpression of HIF-1 alpha and HIF-2 alpha in a primary
neuronal model of hypoxia. J Cereb Blood Flow Metab. 24:245–258.
2004. View Article : Google Scholar : PubMed/NCBI
|