Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most
prevalent subtype of non-Hodgkin's lymphoma that develops in
adults, accounting for ~40% of diagnoses resulting from the
transformation of follicular lymphoma (1). Despite large improvements in
chemotherapy regimens, a substantial percentage of patients (~40%)
succumb to DLBCL, primarily due to drug resistance and disease
relapse (2), thus indicating the
urgent requirement for increased specificity chemotherapy.
MicroRNAs (miRNAs/miRs) are non-coding RNAs that
function to regulate gene expression in a number of cellular
processes, including development, differentiation and proliferation
(3,4).
During the last decade, miRNAs have gained increasing attention
from researchers worldwide due to their crucial functions in human
disease and their potential to serve as therapeutic targets.
Notably, miR-187 in particular has been reported to be associated
with certain types of cancer and other diseases: Zhao et al
(5) reported that an increased level
of miR-187 inhibits tumor invasiveness during the later stages of
carcinogenesis, and Casanova-Salas et al (6) identified that miR-182 and miR-187 may
serve as biomarkers for the prognosis of patients with prostate
cancer by identifying risk of progression. Locke et al
(7) observed that the elevated
expression of miR-187 in pancreatic islets from patients with type
2 diabetes was associated with decreased glucose-stimulated
secretion of insulin. However, the expression pattern and functions
of miR-187 in DLBCL cells has not been identified. Further
investigation into miR-187 as a novel therapeutic target may aid
the development of a successful therapeutic strategy for patients
with DLBCL.
Studies have described B-cell lymphoma 6 (BCL6) as a
key regulator of B lymphocyte growth and development (8,9), with
modified BCL6 expression implicated in the pathogenesis of DLBCL
(10–12). The majority of DLBCL cells maintain a
high expression level of BCL6, but the underlying mechanisms that
regulate this are not sufficiently understood. In the present
study, the association between miR-187 and BCL6 was investigated,
alongside the functions of miR-187 in DLBCL cell apoptosis and
multidrug resistance.
Materials and methods
Cell culture, plasmid construction and
transfection
The human DLBCL cell lines SUDHL2 and OCI-LY3 and
the Burkitt's lymphoma cell line Raji (purchased from Type Culture
Collection of the Chinese Academy of Sciences, Shanghai, China)
were cultured in RPMI 1640 medium containing 10% fetal bovine
serum. The cells were incubated at 37°C in a humidified atmosphere
of 5% CO2 in air. Healthy B cells were obtained from The
Cell Bank of Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). miR-Report BCL6 3′-untranslated regions (UTRs)
is the predicted miR-187 binding sites, which were commercially
constructed by Guangzhou RiboBio Co., Ltd. (Guangzhou, China), and
mutation of the potential miR-187 binding sites on the miR-Report
BCL6 3′-UTR was performed by Beijing Transgen Biotech Co., Ltd.
(Beijing, China). The pcDNA3-BCL6 overexpression plasmid was
constructed by GeneChem Co., Ltd. (Shanghai, China), and pcDNA3 was
used as the empty vector for control. The scramble and miR-187
mimics were purchased from RiboBio Co., Ltd. The miR-187 mimics are
synthesized fragments that share the same sequence as miR-187. The
scramble miR was used as a negative control. Transfection was
performed using Gene Pulser Xcell™ Electroporation system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) according to the
manufacturer's protocols. The medium was changed with fresh culture
medium at 6–8 h post transfection.
Reverse transcription
(RT)-quantitative polymerase chain reaction (qPCR)
RNA was extracted from the healthy B cells and Raji,
OCI-Ly3 and SUGHL2 cells lines using TRIzol® Reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) based on the manufacturer's
protocols. cDNA was synthesized from 2 µg total RNA using the M-MLV
Reverse Transcriptase (Promega Corporation) in a 20-µl reaction
mixture. RT-qPCR was performed using the Applied Biosystems 7300
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.), with the SYBR® Green Realtime PCR Master mix (Toyobo Co.,
Ltd., Osaka, Japan) and the appropriate primers. The cDNA was
denatured at 95°C for 3 min, and subsequently amplification and
fluorescence determination were performed in three steps:
Denaturation at 95°C for 15 sec; annealing at 56°C for 20 sec; and
extension at 72°C for 20 sec. The temperature was decreased to 50°C
and raised slowly to 95°C using a temperature transition rate of
0.1°C/sec. The detection of SYBR Green fluorescence, which reflects
the amount of double-stranded DNA, was performed at the process of
annealing. The amplification cycle number was 45 for all target
genes. To discriminate specific from nonspecific PCR products, a
melting curve was obtained at the end of each run. All the data
were the average of at least three independent experiments. The RT
and PCR primers for miR-187 and the endogenous control, U6, were
purchased from Guangzhou RiboBio Co., Ltd as follows: miR-187,
forward 5′-ACACTCCAGCTGGGGGCCGACGTTGTGTT-3′ and reverse
5′-TGGTGTCGTGGAGTCG-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and
reverse 5′-AACGCTTCACGAA-3′. Other primers were also purchased from
Guangzhou RiboBio Co., Ltd and included the following: BCL6 (76
bp), forward 5′-GGAGTCGAGACATCTTGACTGA-3′, reverse
5′-ATGAGGACCGTTTTATGGGCT-3′; 18S (101 bp), forward
5′-ACAACTTTGGTATCGTGGAAGG-3′, reverse 5′-GCCATCACGCCACAGTTTC-3′.
Relative mRNA expression levels were calculated as
2-∆∆Cq (13) and were
normalised against U6.
Western blotting
Cells were homogenized using 1X Cell Lysis buffer
(Cell Signaling Technology, Inc., Danvers, MA, USA) followed by
5–10 min boiling and centrifugation at 12,000 × g (Eppendorf®
Microcentrifuge 5415D; Eppendorf, Hamburg, Germany) to obtain the
supernatant. Samples containing 50 µg of total protein were
separated on a 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis gel and transferred onto nitrocellulose membranes
(Bio-Rad Laboratories, Inc.). Following saturation with 5% (w/v)
non-fat dry milk in Tris-buffered saline and 0.1% (w/v) Tween 20
(TBST; Beijing Biosea Biotechnology Co., Ltd., Beijing, China), the
membranes were incubated with the following antibodies at 4°C
overnight: Rabbit polyclonal anti-caspase-3 (catalog no., sc-7148;
dilution, 1:1,000), rabbit polyclonal anti-cleaved-caspase-3
(catalog no., sc-22171-R; dilution, 1:1,000), mouse monoclonal poly
ADP ribose polymerase (PARP; catalog no., sc-53643; dilution,
1:500), rabbit polyclonal anti-cleaved-PARP (cPARP; catalog no.,
sc-23461-R; dilution, 1:500), mouse monoclonal anti-BCL-6 (catalog
no., sc-56625; dilution, 1:1,000) and goat polyclonal
anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; catalog no.,
sc-48166; dilution, 1:2,000) (Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA); rabbit polyclonal anti-β-actin antibody (catalog
no., AP0733; dilution, 1:5,000; Bioworld Technology, Inc., St.
Louis Park, MN, USA). Following 3 washes with TBST, the membranes
were incubated with donkey anti-mouse immunoglobulin (Ig)G
conjugated to horseradish peroxidase (HRP) (catalog no., sc-2314;
dilution, 1:10,000), donkey anti-goat IgG-HRP (catalog no.,
sc-2020; dilution, 1:20,000) and goat anti-rabbit IgG-HRP secondary
antibodies (catalog no., sc-2004; dilution, 1:5,000)(Santa Cruz
Biotechnology, Inc.) conjugated to IRDye® 800CW Infrared Dye
(LI-COR Biotechnology, Lincoln, NE, USA). Following 1 h of
incubation at 37°C, the membranes were washed 3 times with TBST.
The blots were visualized using the Odyssey® CLx Infrared Imaging
system (LI-COR Biotechnology). The signals were densitometrically
assessed (Odyssey Application software; version 3.0) and normalized
to β-actin or GAPDH.
Luciferase assays
For the overexpression or suppression of miR-187,
100 pM miR-187 mimics or inhibitors were introduced into
1×106 SUDHL2 cells via electroporation (1,600 V, 20 Ω, 1
pulse; ECM 830 Square Wave Electroporation System; BTX® Harvard
Apparatus, Inc., Holliston, MA, USA). For the luciferase assay, 2
µg miR-Report BCL6 3′-UTR plasmid or the vector control were
transferred into 1×106 SUDHL2 cells using
electroporation according to the manufacturer's protocols.
Following cell transfection for 24 h, the cells were lysed, and
luciferase activities was measured using a Dual-Luciferase®
Reporter Assay system (Promega Corporation), according to the
manufacturer's protocol. Luciferase activity was measured and
normalized to Renilla luciferase activity.
Flow cytometry analysis
Following appropriate treatment, the cultured cells
were harvested and washed with phosphate-buffered saline. For each
sample, 5×105 cells were stained with the Annexin
V-FITC/PI Staining Detection kit (Beijing Biosea Biotechnology Co.,
Ltd.) for 30 min at room temperature, and were subsequently
analyzed using flow cytometry (FACSCanto II; BD Biosciences,
Franklin Lakes, NJ, USA).
MTT assay and cell counting kit 8
(CCK-8) assay
Doxorubicin (DOX), vincristine (VCR) and bortezomib
were purchased from Sigma-Aldrich (St. Louis, MO, USA). The SUDHL2
cells were treated with the drugs at concentrations ranging between
0 and 250 nM (0, 50, 100, 150, 200 and 250 nM) for 48 h; the medium
was then discarded, and 20 µl 0.05% MTT was added and subsequently
incubated for 4 h. The medium was aspirated following
centrifugation (7,200 × g), and dimethyl sulfoxide was added and
mixed for 2 min. The IC50 for each drug was detected by
a microplate reader (SpectraMax® M5 Microplate Reader; Molecular
Devices, LLC, Sunnyvale, CA, USA). For CCK-8 assay, cells were
seeded in 96-well plates at a density of 2,000 cells per well. The
absorptions of the cells were measured using a CCK-8 kit (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan), according to the
manufacturer's protocols at different indicated time points.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Comparisons between groups were analyzed using Student's t
test or analysis of variance, and the Student-Newman-Keuls method
was utilized to estimate the level of significance. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression pattern and function of
miR-187 in B-cell lymphoma cells
To determine whether miR-187 functions in lymphoma
development, the expression pattern of miR-187 was analyzed by
RT-qPCR in various types of lymphoma cells (SUDHL2, OCI-LY3 and
Raji cells). B cells isolated from the healthy donors were used as
a control. The results revealed that miR-187 expression was
significantly lower in the B-cell lymphoma cells compared with the
healthy B cell controls. However, expression patterns appeared to
vary depending on the cell type, miR-187 expression levels in the
SUDHL2 and OCI-LY3 cells were 7.3 and 16.1%, respectively, of those
observed in the healthy B cells (P<0.01), whilst the Raji cell
miR-187 expression was 57.6% of that observed in the healthy B
cells (P<0.05) (Fig. 1A).
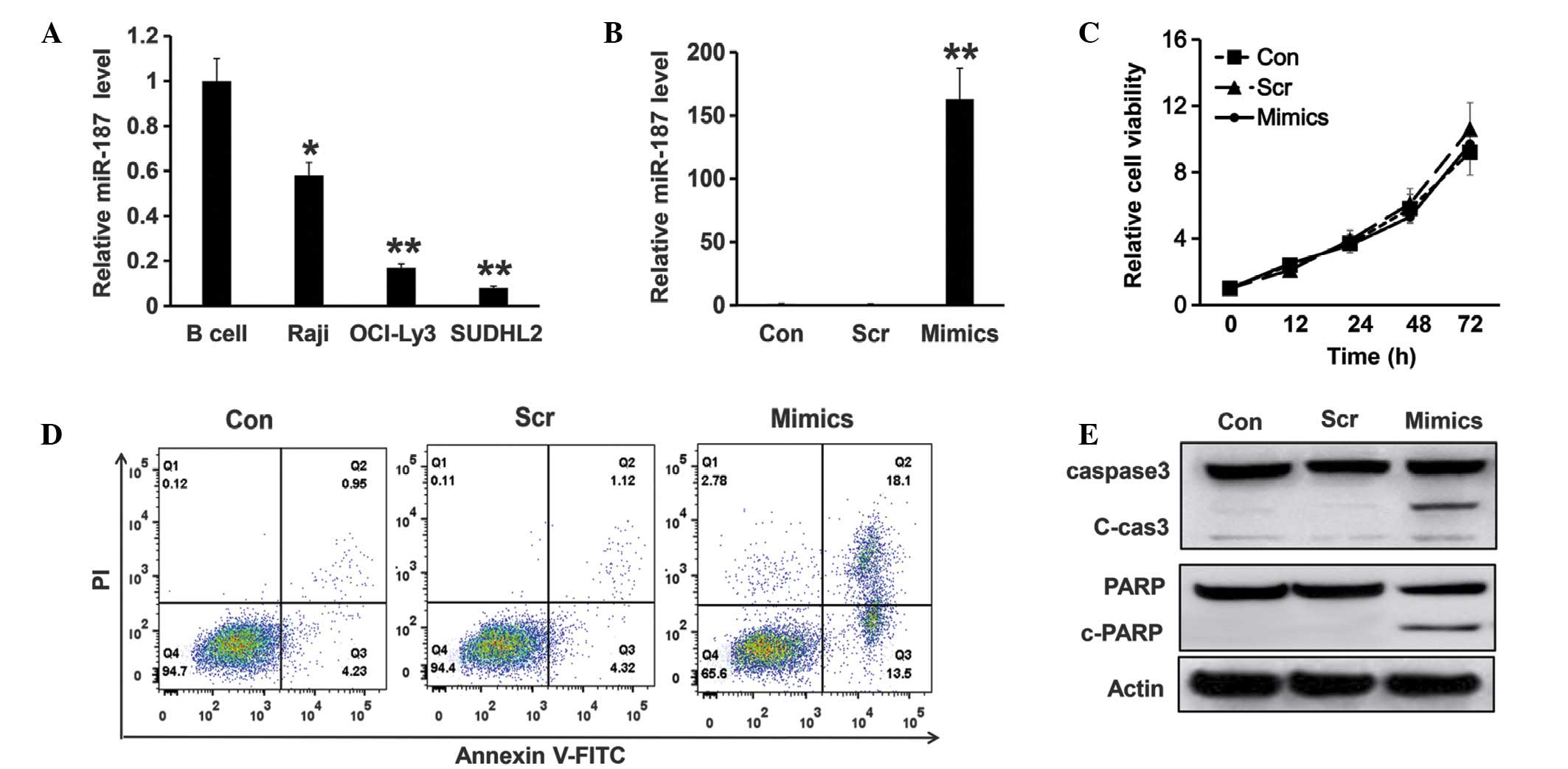 | Figure 1.Expression and function of miR-187 in
diffuse large B-cell lymphoma cells. (A) Relative expression level
of miR-187 was detected via reverse transcription-quantitative
polymerase chain reaction in B-cells from healthy donors, Raji
cells, OCI-LY3 and SUDHL cells. The U6 gene was used as an
endogenous control. (B) Relative expression of miR-187 in Con,
SUDHL cells electroporated with 100 pM mimics or Scr negative
control for 24 h. All experiments were repeated at least three
times. (C) Cell viability was analyzed using CCK-8 assay in the
SUDHL cells transfected with scramble or miR-187 mimics compared
with the parental control. (D) Flow cytometry assay using annexin
V-FITC and PI staining was utilized to detect the apoptosis of the
Con cells, SUDHL cells electroporated with 100 pM mimics or Scr
negative control for 48 h. (E) Western blot analysis was used to
detect the cleavage activity of caspase-3 and PARP in the SUDHL2
cells with different treatment for 48 h. *P<0.05 and **P<0.01
vs. controls. miR, microRNA; Con, parental control SUDHL cells;
Scr, scramble; Mimics, miR-187 mimics; FITC, fluorescein
isothiocyanate; PI, propidium iodide; PARP, poly ADP ribose
polymerase. |
To study the function of miR-187 in DLBCL cells, its
expression was induced using the miR-187 mimics or the scramble
control in the SUDHL2 cells via electroporation; RT-qPCR was then
utilized to detect miR-187 levels. As shown in Fig. 1B, miR-187 was significantly
upregulated by the miR-187 mimics at 24 h post electroporation
(P<0.01) compared to the controls. To analyze whether miR-187
serves a role in the cell growth of SUDHL2 cells, a CCK-8 assay was
performed to assess the proliferation of the parental control
SUDHL2 cells and SUDHL2 cells overexpressing miR-187 mimics or
scramble control. The results of CCK-8 assay demonstrated that
there was no significant difference between the proliferation of
the parental control and the miR-187 overexpressing cells (Fig. 1C). However, miR-187 overexpression did
affect apoptosis in SUDHL2 cells. SUDHL2 cells that had
overexpressed the miR-187 mimic for 48 h were subjected to flow
cytometric analysis to assess the number of apoptotic cells in each
treatment group. It was observed that miR-187 overexpression
resulted in an increased percentage of apoptotic cells compared
with the scramble and parental control groups (Fig. 1D). This result was also verified by
western blot analysis, which demonstrated that the cleavage
activity of caspase-3 and PARP significantly increased in the
SUDHL2 cells overexpressing miR-187 compared with the control
groups (Fig. 1E).
miR-187 enhances the chemotherapeutic
sensitivity of SUDHL2 cells
Drug resistance is a major challenged faced during
the treatment of lymphoma (14,15). To
evaluate whether miR-187 serves a role in the drug resistance of
DLBCL cells, the chemotherapeutic sensitivity of the parental
SUDHL2 cells, or SUDHL2 cells overexpressing miR-187 or the
scramble control, to 5 drugs used clinically was analyzed via MTT
assay. The drugs included DOX, VCR and bortezomib, which were used
in testing the chemotherapeutic sensitivity of the cells. The
results demonstrated that, following transfection of the miR-187
mimics and scramble control, the capability of multidrug resistance
in the SUDHL2 cells. Overexpressing miR-187 significantly decreased
the concentrations of DOX between 78±8.5 and 46±5.4 (P<0.05),
bortezomib between 52±2.1 and 46±4.9 (P<0.01) and VCR between
73±2.5 and 42±6.1 (P<0.05), suggesting that miR-187 serves an
important role in multidrug resistance (Table I).
 | Table I.Comparison of IC50 of each
group for different drugs. |
Table I.
Comparison of IC50 of each
group for different drugs.
|
| IC50,
nmol/l (mean ± SD) |
|---|
|
|
|
|---|
| Drug | Control | Scramble | Mimics |
|---|
| Doxorubicin | 78±8.5 | 82±10.1 | 46±5.4a |
| Bortezomib | 52±2.1 | 55±8.4 | 46±4.9b |
| Vincristine | 73±2.5 | 69±10.4 |
42±6.1a |
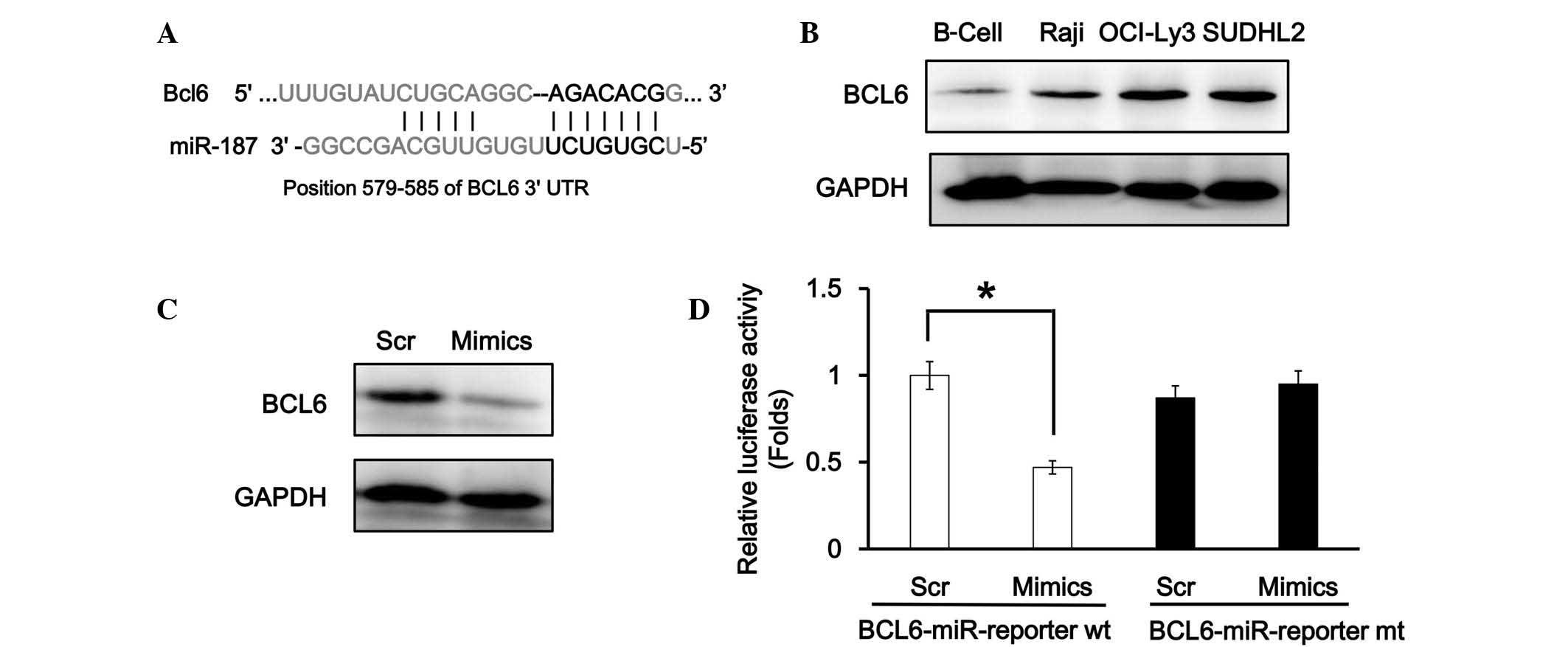
BCL6 is a target gene of miR-187
To elucidate the detailed molecular mechanisms of
miR-187 in DLBCL cells, its potential targets were evaluated using
TargetScan (www.targetscan.org), which indicated
that there is one miR-187 conserved binding site in the BCL6 3′-UTR
(Fig. 2A). In the DLBCL cell lines,
western blot analysis revealed that BCL6 was significantly
upregulated in the SUDHL2 and OCI-LY3 cells, and slightly
upregulated in the Raji cells, whereas there was limited BCL6
expression detected in the B cell control; this indicated a
negative association with miR-187 (Fig.
2B). In addition, western blot analysis indicated that the
overexpression of miR-187 attenuated BCL6 expression in the SUDHL2
cells (Fig. 2C). The miR-Report
luciferase assay was performed to determine whether miR-187
directly targeted the 3′-UTR of BCL6. The luciferase assay
indicated that miR-187 overexpression significantly reduced the
luciferase activity in the wild-type BCL6 3′-UTR reporter
(P>0.05), but not in the mutant reporter (Fig. 2D).
Exogenous BCL6 reverses miR-187
function
In order to determine whether miR-187 regulates
apoptosis and multidrug resistance through its target BCL6, miR-187
mimics and BCL6 overexpression plasmids were co-transfected into
the SUDHL2 cells. Western blot analysis indicated that the BCL6
downregulation induced by miR-187 was completely reversed by
exogenous BCL6, as the BCL6 overexpression plasmid lacked miR-187
binding sites (Fig. 3A).
Subsequently, flow cytometry was performed, and the results
demonstrated that, although there was abundant miR-187 in the DLBCL
cells, BCL6 overexpression continued to inhibit cell apoptosis
(Fig. 3B and C). Furthermore, the
suppression of multidrug resistance capability induced by miR-187
was significantly reversed following BCL6 overexpression (Table II). In the absence of the miR-187 and
BCL6 3′-UTR interaction, the effect of miR-187 was diminished, thus
suggesting that miR-187 mediates apoptosis and multidrug resistance
through its transcriptional modulation of BCL6.
 | Figure 3.Exogenous expression of BCL6 in the
SUDHL2 cells reversed the miR-187 mimic function. (A) Western
blotting was utilized to detect the protein level of BCL6 in the
miR-187 overexpressing SUDHL cells transfected with BCL6 OE or a
vector control (vector). (B) Percentage of apoptotic cells was
counted in miR-187 overexpressing SUDHL cells transfected with BCL6
OE or a vector control. All experiments were repeated at least
three times. **P<0.01 vs. controls. (C) A flow cytometry assay,
using annexin V-FITC and PI staining, was performed to detect the
apoptotic cells co-transfected in the miR-187 overexpressing SUDHL
cells transfected with BCL6 OE or vector control (vector). BCL6 OE,
BCL6 overexpressing plasmid; BCL6, B-cell lymphoma 6; miR,
microRNA; Annexin V-FITC, Annexin V-fluorescein isothiocyanate; PI,
propidium iodide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase;
Comp-PI-A, propidium iodide; Comp-FITC-A, Annexin V-fluorescein
isothiocyanate. |
 | Table II.Comparison of IC50 of each
group for different drugs. |
Table II.
Comparison of IC50 of each
group for different drugs.
|
| IC50,
nmol/l (mean ± SD) |
|---|
|
|
|
|---|
|
|
| miR-187
overexpression |
|---|
|
|
|
|
|---|
| Drug | Control | Vector | BCL6 OE |
|---|
| Doxorubicin | 82±2.1 | 79±6.2 | 129±13.9a |
| Bortezomib | 42±2.14 | 48±3.5 | 96±4.9b |
| Vincristine | 68±5.8 | 71±6.3 | 157±12.1a |
Discussion
Over the past decade, miRNAs have gained increasing
attention from researchers regarding their possible role in cancer
biology, with a number of miRNAs being demonstrated to serve
crucial regulatory roles in cancer development and tumorigenesis.
Based on the accumulating data, it has been debated as to whether
miRNAs may serve as novel therapeutic targets for numerous
malignancies (16).
Research regarding the biological function of
miR-187 in carcinogenesis remains in its infancy. Studies have
implicated miR-187 activity in the development of various types of
tumors (5,17,18);
however, its role in DLBCL has not yet been established.
The BCL6 protein, a zinc finger transcription factor
that has been conserved throughout evolution, has demonstrated high
expression levels in numerous types of human cancer, alongside
lymphoid system malignancies (19).
Further studies have observed potent BCL6 protein expression in
various types of human cancer outside of the lymphoid system. For
example, overexpression of BCL6 protein was demonstrated in breast
cancer tissues, particularly in high-grade ductal breast cancer,
when compared to normal mammary gland tissues (20,21).
Overall, the specific function of BCL6 protein in carcinogenesis
remains to be clarified.
The majority of germinal center B-cell-like (GCB)
and activated B-cell-like type DLBCL cells are dependent upon BCL6
in order to maintain their proliferation and survival, reflecting
the function of BCL6 in GCB DLBCL and supporting the notion that it
is a broadly relevant therapeutic target in DLBCL (22–24).
The effect of miR-187 on apoptosis and multidrug
resistance in DLBCL cells is most likely achieved through BCL6
regulation. The following evidence supports this hypothesis: i) The
expression of miR-187 is negatively correlated with BCL6 expression
in DLBCL cell lines; and ii) BCL6 is a target of miR-187, with
miR-187 indirectly promoting lymphoma cell apoptosis and enhancing
multidrug sensitivity by targeting its 3′-UTR.
To the best of our knowledge, there are no studies
currently published that report of a direct involvement of miR-187
and BCL6 in DLBCL carcinogenesis, thus warranting further
investigation of their possible underlying mechanisms in DLBCL. The
results of the present study provide evidence that miRNAs may be
useful for future development of novel therapeutic strategies for
DLBCL. Further studies, possibly utilizing animal models, alongside
additional human samples, are required to validate the results from
the present study, aiming to further elucidate the functions of
miRNA and aid the treatment of lymphoma.
References
|
1
|
Sabattini E, Bacci F, Sagramoso C and
Pileri SA: WHO classification of tumours of haematopoietic and
lymphoid tissues in 2008: An overview. Pathologica. 102:83–87.
2010.PubMed/NCBI
|
|
2
|
Lenz G and Staudt LM: Aggressive
lymphomas. N Engl J Med. 362:1417–1429. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou C, Lu Y and Li X: miR-339-3p inhibits
proliferation and metastasis of colorectal cancer. Oncol Lett.
10:2842–2848. 2015.PubMed/NCBI
|
|
4
|
Vicente R, Noël D, Pers YM, Apparailly F
and Jorgensen C: Deregulation and therapeutic potential of
microRNAs in arthritic diseases. Nat Rev Rheumatol. 2015.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y,
Fan S and Liu Y: MicroRNA-187, down-regulated in clear cell renal
cell carcinoma and associated with lower survival, inhibits cell
growth and migration though targeting B7-H3. Biochem Biophys Res
Commun. 438:439–444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Casanova-Salas I, Rubio-Briones J,
Calatrava A, Mancarella C, Masiá E, Casanova J, Fernández-Serra A,
Rubio L, Ramírez-Backhaus M, Armiñán A, et al: Identification of
miR-187 and miR-182 as biomarkers of early diagnosis and prognosis
in patients with prostate cancer treated with radical
prostatectomy. J Urol. 192:252–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Locke JM, da Silva Xavier G, Dawe HR,
Rutter GA and Harries LW: Increased expression of miR-187 in human
islets from individuals with type 2 diabetes is associated with
reduced glucose-stimulated insulin secretion. Diabetologia.
57:122–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang CC, Ye BH, Chaganti RS and
Dalla-Favera R: BCL-6, a POZ/zinc-finger protein, is a
sequence-specific transcriptional repressor. Proc Natl Acad Sci
USA. 93:6947–6952. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jardin F, Ruminy P, Bastard C and Tilly H:
The BCL6 proto-oncogene: A leading role during germinal center
development and lymphomagenesis. Pathol Biol (Paris). 55:73–83.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Duan S, Cermak L, Pagan JK, Rossi M,
Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R and Pagano
M: FBXO11 targets BCL6 for degradation and is inactivated in
diffuse large B-cell lymphomas. Nature. 481:90–93. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pasqualucci L, Bereshchenko O, Niu H,
Klein U, Basso K, Guglielmino R, Cattoretti G and Dalla-Favera R:
Molecular pathogenesis of non-Hodgkin's lymphoma: The role of
Bcl-6. Leuk Lymphoma. 44(Suppl 3): S5–S12. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ye BH: BCL-6 in the pathogenesis of
non-Hodgkin's lymphoma. Cancer Invest. 18:356–365. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Anstaett OL, Brownlie J, Collins ME and
Thomas CJ: Validation of endogenous reference genes for RT-qPCR
normalisation in bovine lymphoid cells (BL-3) infected with Bovine
Viral Diarrhoea Virus (BVDV). Vet Immunol Immunopathol.
137:201–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang N, Chen W, Zhang JW, Li Y, Zeng XC,
Zhang T, Fu BS, Yi HM and Zhang Q: Aberrantly regulated dysadherin
and B-cell lymphoma 2/B-cell lymphoma 2-associated X enhances
tumorigenesis and DNA targeting drug resistance of liver cancer
stem cells. Mol Med Rep. 12:7239–7246. 2015.PubMed/NCBI
|
|
15
|
Camicia R, Winkler HC and Hassa PO: Novel
drug targets for personalized precision medicine in
relapsed/refractory diffuse large B-cell lymphoma: A comprehensive
review. Mol Cancer. 14:2072015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Soriano A, Jubierre L, Almazán-Moga A,
Molist C, Roma J, de Toledo JS, Gallego S and Segura MF: MicroRNAs
as pharmacological targets in cancer. Pharmacol Res. 75:3–14. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mulrane L, Madden SF, Brennan DJ, Gremel
G, McGee SF, McNally S, Martin F, Crown JP, Jirström K, Higgins DG,
et al: miR-187 is an independent prognostic factor in breast cancer
and confers increased invasive potential in vitro. Clin
Cancer Res. 18:6702–6713. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chao A, Lin CY, Lee YS, Tsai CL, Wei PC,
Hsueh S, Wu TI, Tsai CN, Wang CJ, Chao AS, et al: Regulation of
ovarian cancer progression by microRNA-187 through targeting
Disabled homolog-2. Oncogene. 31:764–775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Q, Liu X, Yan H, He YH, Ye S, Cheng XW,
Zhu GL, Wu WY, Wang XN, Kong XJ, et al: B-cell lymphoma 6 protein
stimulates oncogenicity of human breast cancer cells. BMC Cancer.
14:4182014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bajalica-Lagercrantz S, Piehl F, Farnebo
F, Larsson C and Lagercrantz J: Expression of the BCL6 gene in the
pre- and postnatal mouse. Biochem Biophys Res Commun. 247:357–360.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bos R, van Diest PJ, van der Groep P,
Greijer AE, Hermsen MA, Heijnen I, Meijer GA, Baak JP, Pinedo HM,
van der Wall E and Shvarts A: Protein expression of B-cell lymphoma
gene 6 (BCL-6) in invasive breast cancer is associated with cyclin
D1 and hypoxia-inducible factor-1alpha (HIF-1alpha). Oncogene.
22:8948–8951. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Polo JM, Dell'Oso T, Ranuncolo SM,
Cerchietti L, Beck D, Da Silva GF, Prive GG, Licht JD and Melnick
A: Specific peptide interference reveals BCL6 transcriptional and
oncogenic mechanisms in B-cell lymphoma cells. Nat Med.
10:1329–1335. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Polo JM, Juszczynski P, Monti S,
Cerchietti L, Ye K, Greally JM, Shipp M and Melnick A:
Transcriptional signature with differential expression of BCL6
target genes accurately identifies BCL6-dependent diffuse large B
cell lymphomas. Proc Natl Acad Sci USA. 104:3207–3212. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cerchietti LC, Yang SN, Shaknovich R,
Hatzi K, Polo JM, Chadburn A, Dowdy SF and Melnick A: A
peptomimetic inhibitor of BCL6 with potent antilymphoma effects in
vitro and in vivo. Blood. 113:3397–3405. 2009. View Article : Google Scholar : PubMed/NCBI
|