Introduction
Ovarian cancer is the most lethal gynecological
malignancy, and <40% of patients with ovarian cancer are cured.
In 2015, it is estimated that 21,290 new cases and 14,180
mortalities will occur in association with this disease in the
United States (1). Epidemiological
studies have identified several risk factors of ovarian cancer,
including nulliparity, hormone therapy and pelvic inflammatory
disease (2–4). Initial treatment for ovarian cancer
consists of surgical staging and cytoreduction, followed by
systemic chemotherapy in the majority of patients. Given the high
mortality rate of ovarian cancer, the identification of novel
biomarkers and therapeutic targets has been a major research focus
for a number of years.
Long non-coding RNA (lncRNA) is an RNA molecule that
is >200 nucleotides long and is not translated into a protein.
Although the current understanding of the role of lncRNAs is
limited, increasing numbers of studies have indicated that lncRNAs
may regulate gene expression at various levels, including chromatin
modification, transcription and posttranscriptional processing
(5–7).
lncRNAs have been reported to control various cellular processes,
including proliferation, apoptosis and invasion, and are implicated
in human diseases, including various types of tumors.
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1), also known as non-coding nuclear-enriched abundant
transcript 2, is a 8,000 nucleotide-long, spliced non-coding RNA,
which is highly conserved in mammals. MALAT1 has been reported to
be deregulated in several tumors, including non-small cell lung
cancer, hepatocellular carcinoma, cervical cancer, bladder cancer
and colorectal cancer (8–12). However, the role of MALAT1 in ovarian
cancer has not been previously investigated.
The present study indicated that MALAT1 inhibition
significantly suppressed tumorigenicity and induced apoptosis in
SKOV3 cells. In addition, the knockdown of MALAT1 was indicated to
alter the expression of a number of genes associated with cell
proliferation, metastasis and apoptosis. To the best of our
knowledge, the present study is the first to report an oncogenic
role for MALAT1 in ovarian cancer.
Materials and methods
Cell lines and cell culture
The human ovarian cancer SKOV3 cell line and 293T
cells were obtained from the American Type Culture Collection
(Manassas, VA, USA). The two cell lines were cultured in Dulbecco's
modified Eagle's medium (HyClone; GE Healthcare Life Sciences,
Logan, UT, USA), supplemented with 10% fetal bovine serum (HyClone;
GE Healthcare Life Sciences), 100 U/ml penicillin and 100 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) in standard culture conditions of 5% CO2 at
37°C.
Establishment of stable
MALAT1-knockdown (KD) and negative control (NC) cells
Four microRNA (miRNA) oligonucleotides targeting
MALAT1 (KD1, KD2, KD3 and KD4; Table
I) were synthesized and inserted into the pcDNA6.2-GW/EmGFP-miR
vector (Invitrogen; Thermo Fisher Scientific, Inc.) and used for
transient transfections. The pcDNA6.2-GW/EmGFP-miR-Neg vector
(Invitrogen; Thermo Fisher Scientific, Inc.) was used as negative
control. Cells were maintained in 5% CO2 at 37°C and
harvested at 2 days post-infection. Reverse
transcription-quantitative polymerase chain reactive (RT-qPCR) was
performed in order to determine the efficiency of MALAT1 knockdown
and to screen for the miRNA with the greatest knockdown efficiency,
which was then used for subsequent experiments.
pLenti6.3-MCS/V5DEST lentiviral vector was purchased from Shanghai
R&S Biotechnology Co., Ltd. (Shanghai, China). The
pLenti6.3-EmGFP-MALAT1-miR and plenti-EmGFP-miR-Neg vectors were
constructed using the pcDNA6.2-GW/EmGFP-miR and
pcDNA6.2-GW/EmGFP-miR-Neg vectors with the greatest knockdown
efficiency, respectively. To produce recombinant lentiviruses, the
293T cells were co-transfected with lentivirus expression plasmids
and packaging plasmid mix (Invitrogen; Thermo Fisher Scientific,
Inc.) using POLOdeliverer™ 3000 Transfection Reagent (Ruisai Inc.,
Shanghai, China). The SKOV3 cells were then infected with these
lentiviruses at a multiplicity of infection of 30, and selected
with blasticidin (Invitrogen; Thermo Fisher Scientific, Inc.). At 2
weeks post-infection, cells were harvested and qRT-PCR was
performed to determine the expression levels of MALAT1 in KD and NC
cells.
 | Table I.Sequences of the four microRNA
oligonucleotides. |
Table I.
Sequences of the four microRNA
oligonucleotides.
| Oligonucleotide | Sequence |
|---|
| KD1 |
|
|
Forward |
5′-TGCTGTCCACTTGATCCCAACTCATCGTTTTGGCCACTGACTGACGATGAGTTGATCAAGTGGA-3′ |
|
Reverse |
5-CCTGTCCACTTGATCAACTCATCGTCAGTCAGTGGCCAAAACGATGAGTTGGGATCAAGTGGAC-3′ |
| KD2 |
|
|
Forward |
5′-TGCTGCTTATCTGTTAACAGCTGCCTGTTTTGGCCACTGACTGACAGGCAGCTTAACAGATAAG-3′ |
|
Reverse |
5′-CCTGCTTATCTGTTAAGCTGCCTGTCAGTCAGTGGCCAAAACAGGCAGCTGTTAACAGATAAGC-3′ |
| KD3 |
|
|
Forward |
5′-TGCTGTGTACTATCCCATCACTGAAGGTTTTGGCCACTGACTGACCTTCAGTGGGGATAGTACA-3′ |
|
Reverse |
5′-CCTGTGTACTATCCCCACTGAAGGTCAGTCAGTGGCCAAAACCTTCAGTGATGGGATAGTACAC-3′ |
| KD4 |
|
|
Forward |
5′-TGCTGTTCCTTAGTTGGCATCAAGGCGTTTTGGCCACTGACTGACGCCTTGATCAACTAAGGAA-3′ |
|
Reverse |
5′-CCTGTTCCTTAGTTGATCAAGGCGTCAGTCAGTGGCCAAAACGCCTTGATGCCAACTAAGGAAC-3′ |
RNA extraction and RT-qPCR
Total RNA was extracted from the cell samples using
the Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. The RNA integrity was
analyzed using the Agilent 2100 Bioanalyzer (Agilent Technologies
Inc., Santa Clara, CA, USA). Each RT reaction consisted of 0.5 µg
RNA, 2 µl PrimerScript Buffer, 0.5 µl oligo(dT), 0.5 µl random
primers and 0.5 µl PrimerScript RT Enzyme mix I (Takara, Otsu,
Japan), in a total volume of 10 µl. Reactions were performed in a
GeneAmp® PCR System 9700 (Applied Biosystems; Thermo
Fisher Scientific, Inc.) for 15 min at 37°C, followed by heat
inactivation of reverse transcriptase for 5 sec at 85°C. The 10-µl
RT reaction mixture was then diluted 10 times in nuclease-free
water and held at −20°C. qPCR was performed using the
LightCycler® 480 II Real-Time PCR instrument (Roche
Diagnostics, Basel, Switzerland) with a total volume including 1 µl
cDNA, 5 µl 2X LightCycler® 480 SYBR Green I Master
(Roche Diagnostics), 0.2 µl of forward primer, 0.2 µl of reverse
primer and 3.6 µl of nuclease-free water. Reactions were incubated
in a 384-well optical plate (Roche Diagnostics) at 95°C for 10 min,
followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec. At
the end of the PCR cycles, melting curve analysis was performed to
validate the specific generation of the expected PCR product. Three
independent experiments were performed, with each sample run in
triplicate. The primers were synthesized by Generay Biotech Co.,
Ltd. (Shanghai, China) and are listed in Table II. The expression levels of the genes
were normalized with regard to GAPDH and were calculated using the
2−ΔΔCq method (13).
 | Table II.Primers used in reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primers used in reverse
transcription-quantitative polymerase chain reaction.
| Gene symbol | Forward primer | Reverse primer |
|---|
| MALAT1 |
ATCAGACCACCACAGGTTTACAG |
GACCATCCCAAAATGCTTCA |
| WISP2 |
GCTTTCTCTCCGACTTCC |
TCTGTGTGCCTTCTCTTCA |
| MUC16 |
ATCATCAGCTATGACATCGAC |
TCTTTCTCAGTGGATAGGCTTA |
| ZEB2 |
TTGGTGTACCAAGAGGCAA |
GCTAAGCCTTCAGTCTGAAT |
| MMP2 |
ACAGAAGGACTCAGGTTG |
GTGGAGAAGAGACTCGGT |
| MMP11 |
CTGAGCAACTGGGCTGTA |
CTGGATTCTGGAGAACAGATTT |
| VEGFA |
GGGCAAATATGACCCAGT |
TGTACCTGTGATCTGTCTTTCT |
| PGF |
CCTTGGAGGAGAGAGACC |
CAGGGAAACAGTTGGCTAA |
| H19 |
TTTCATCCTTCTGTCTCTTTGT |
CAACCAGTGCAAATGACTTAG |
| ERBB4 |
AATAAATGCTAAACCAAGCGT |
TAACAACTAGAAGCTGATGCAC |
| SNAI2 |
TGCAGACCCATTCTGATGTAA |
GCGTCACTCAGTGTGCTA |
| E2F8 |
CCCAAAGGGTCACAATTAGT |
GGGACAAAGAGAGTTCCTAAG |
| CDK2 |
CCCTTGTTTGTCCCTTCTAC |
CATCTCTCACCTGCCTCATA |
| CDC20 |
TGGATCAAAGAGGGCAACT |
GTGAACCACTGGACAGGATA |
| ADRA1B |
AAGAGAACCACCAAGAACCTA |
AAGGGTGTCCTCGTGAAA |
| RASGRP1 |
GTTCCAAACTAAAGCCAATAGC |
ATGGGAACTGATCTTTCATCAA |
| FGF1 |
TTTCAACTTTAGAACCGGGTC |
AAACAGAGCAGGGAACTAC |
| BAX |
AGATGTGGTCTATAATGCGTT |
TCAAGTCAAGGTCACAGTGAG |
| FN1 |
AAGATTCCCGAGAGTAAATCAT |
TCTAAGCTGGGTCTGCTAAC |
| MTBP |
GAAAGCCACAAACAGAACG |
TTGACCTGAACTGGTCTTTGTA |
| ECT2 |
ACAACTCATTTGATATGAAGCG |
AGCTTTCACCAAGTGCTAA |
| GAPDH |
TGTTGCCATCAATGACCCCTT |
CTCCACGACGTACTCAGCG |
Cell proliferation, colony formation,
invasion and motility assays
Cell viability was determined at 24, 48, 72 and 96 h
using the Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto,
Japan), according to the manufacturer's protocol. For the colony
formation assay, cells were counted, seeded at low density (1,000,
1,500 and 2,000 cells/plate), and allowed to grow until visible
colonies appeared. Cells were then stained with crystal violet
(GenMed, Shanghai, China) and colonies were counted by eye. In
vitro invasion assay was performed using 24-well Transwell
units (Corning Life Science, Tewksbury, MA, USA) with polycarbonate
filters coated on the upper side with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). The cells were harvested, and
4×104 cells were placed in the upper part of the
Transwell unit and were allowed to invade the membrane for 72 h at
37°C. Successfully penetrating cells were fixed, stained and
quantified at an optical density of 570 nm. The motility assay was
conducted in a similar fashion without coating with Matrigel. In
total, 8×104 cells were seeded per well and incubated in
5% CO2 at 37°C for 5 h prior to detection. Successfully
penetrating cells were quantified at an optical density of 570 nm
using a microplate reader (Infinite 200 Pro; Tecan Schweiz AG,
Männedorf, Switzerland). Each experiment was performed in
triplicate.
Apoptosis assay
Cell apoptosis was determined by flow cytometric
analysis. Briefly, floating and attached cells were collected,
resuspended, stained with Annexin V-allophycocyanin (BD
Biosciences) and incubated for 30 min in the dark at 20°C. The
analysis was then performed using a BD FACSAria II flow cytometer
(BD Biosciences).
In vivo tumorigenic assay
Mice were housed and maintained in a specific
pathogen-free facility under controlled environmental conditions
(temperature, 22±2°C; humidity, 55±10%) with ad libitum
access to rodent chow and water. KD cells or NC cells
(5×106) were injected subcutaneously into the left
armpit of nude mice (6 to 7 weeks old; n=7/group). Once palpable
tumors developed, caliper measurements were taken twice a week and
tumor volume was calculated on the basis of width (x) and length
(y): x2y/2, where x<y. All mice were sacrificed by
cervical dislocation when tumors reached 2 cm3 in size,
and tumors were collected.
Microarray analysis and verification
of selected genes by RT-qPCR
Microarray analysis was performed by Shanghai
OeBiotech. Co., Ltd. (Shanghai, China), using SurePrint G3 Human
Gene Expression 8×60 K version 2 software (Agilent Technologies,
Inc.). Briefly, total RNAs from KD cells and control cells were
used to synthesize complementary DNA, from which labeled
complementary RNA was then synthesized and hybridized to SurePrint
G3 Human Gene Expression 8×60K v2 (Agilent Technologies, Inc.).
Subsequent to hybridization, processed slides were washed using the
Gene Expression Wash Pack (Agilent Technologies, Inc.) and scanned
using the Agilent Microarray Scanner (Agilent Technologies, Inc.).
The acquired array images were analyzed using Agilent Feature
Extraction software (version 10.7; Agilent Technologies, Inc.),
which performs background subtractions. Quantile normalization and
subsequent data processing were performed using the GeneSpringGX
version 11.0 software package (Agilent Technologies, Inc.). A
threshold of a >2-fold change was used to screen upregulated or
downregulated genes. A total of 20 differentially-expressed genes,
which have been reported to be associated with cell proliferation,
metastasis and apoptosis, were subsequently selected for validation
using RT-qPCR.
Statistical analyses
All statistical analyses were performed using the
SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Differences
between means were analyzed using a two-tailed Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
KD and NC cells were successfully
established
The four pcDNA6.2-GW/EmGFP-miR vectors harboring
various miRNAs (KD1, KD2, KD3 and KD4) were used to transiently
infect 293T cells. In parallel, an NC experiment was performed. The
highest knockdown efficiency was achieved using KD3 (71% relative
to the NC group), which was then used for producing recombinant
lentiviruses. The results of the RT-qPCR showed that the expression
of MALAT1 was decreased by 77% in KD cells, compared with NC cells
(P=0.000) (Fig. 1A).
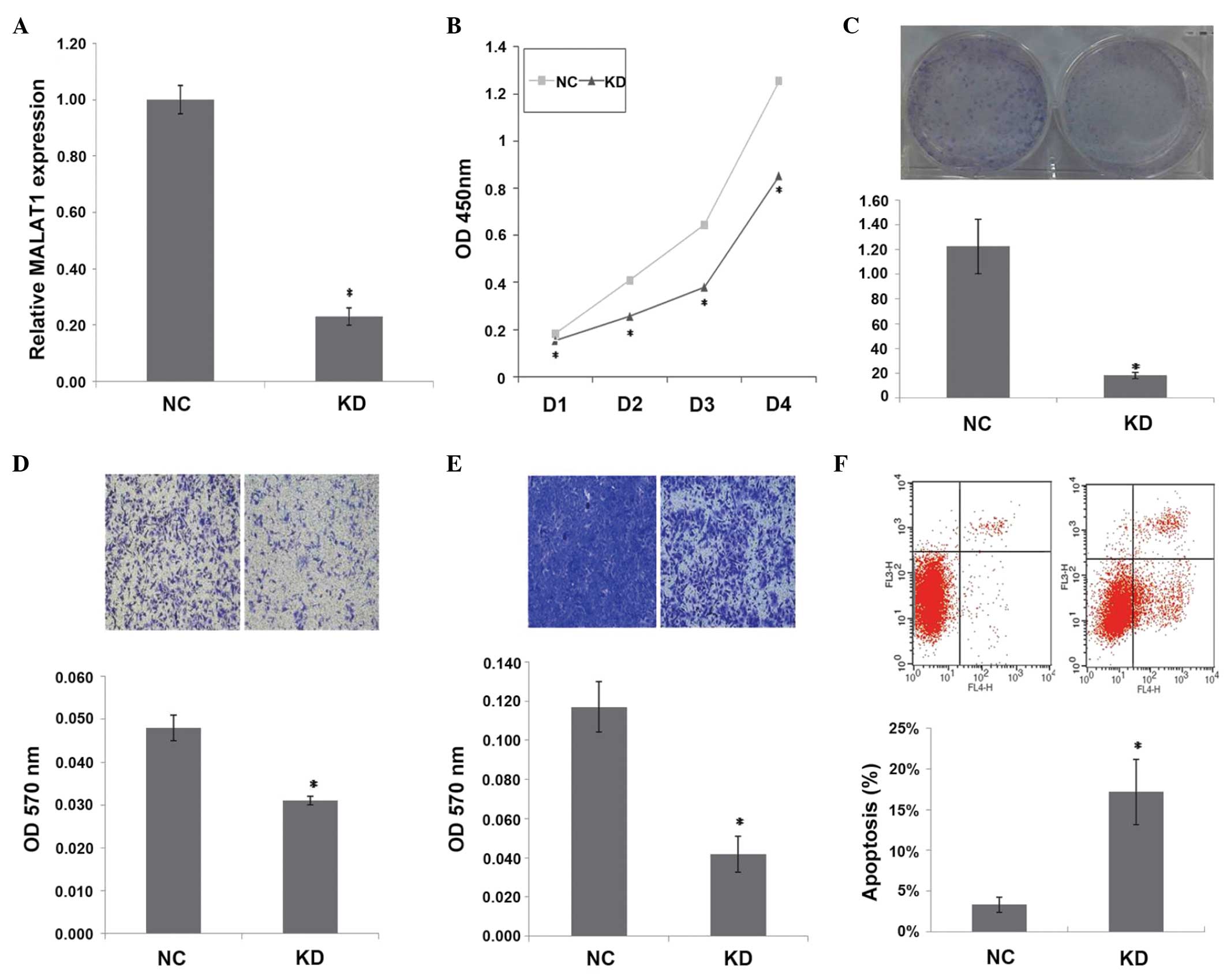 | Figure 1.Inhibition of MALAT1 expression
suppresses tumorigenicity and induces apoptosis in vitro.
(A) Relative MALAT1 expression (*P=0.000), (B) cell proliferation
assay (*P<0.01), (C) colony formation assay (*P=0.001), (D)
Transwell-migration assay (*P=0.002), (E) invasion assay (*P=0.001)
and (F) apoptosis assay (*P=0.004) in KD cells and NC cells.
MALAT1, metastasis-associated lung adenocarcinoma transcript 1; NC,
normal control cells; KD, stable MALAT1-knockdown cells; OD,
optical density; D1, day 1; D2, day 2; D3, day 3; D4, day 4. |
MALAT1 inhibition suppresses
tumorigenicity in vitro
As shown in Fig. 1B, a
significant decrease in cell proliferation was observed over time
in KD cells compared with NC cells (P<0.01). MALAT1 inhibition
also decreased the clonogenicity of KD cells compared with NC cells
(P=0.001) (Fig. 1C). Transwell
migration and invasion assays showed that MALAT1 inhibition
decreased the migration (P=0.002)and invasion (P=0.001) of KD cells
(Fig. 1D and E). These observations
suggest that MALAT1 knockdown suppresses the tumorigenicity of
ovarian cancer cells in vitro.
MALAT1 inhibition induces apoptosis in
KD cells
Apoptosis in KD and NC cells was measured by flow
cytometry analysis. The mean apoptotic cell fractions (early
apoptotic+apoptotic) were significantly increased upon MALAT1
inhibition compared with the negative control (P=0.004), with a
concomitant decrease in the viable cell population (Fig. 1F). This finding suggests an
anti-apoptotic role of MALAT1 in SKOV3 cells.
MALAT1 inhibition suppresses tumor
growth in vivo
Since the in vitro data revealed the
antitumorigenic role of MALAT1 inhibition in ovarian cancer cells,
the effect of MALAT1 inhibition on tumor growth was examined in
vivo. As shown in Fig. 2, the
mean volume of the tumors in the KD group was significantly smaller
compared with the tumors in the NC group (P<0.01), suggesting
that the inhibition of MALAT1 significantly suppressed the
tumorigenicity of SKOV3 cells in vivo.
Genes identified during the knockdown
of MALAT1 expression
In order to additionally characterize the function
of MALAT1 within cells, RNA isolated from the KD cells was
hybridized to the Agilent gene expression microarray and compared
with the RNA isolated from the NC group. Data analysis showed that,
when compared with the NC cells, a >2-fold deregulation was
indicated in 921 genes in the KD cells. Since MALAT1 inhibition
suppresses tumorigenicity and induces apoptosis in ovarian cancer
cells, 20 genes, which have previously been reported to be
associated with cell proliferation, metastasis and apoptosis, were
selected for validation. As shown in Table III, 19 of these genes confirmed the
deregulation that was indicated by the microarray analysis.
 | Table III.Deregulated genes in
metastasis-associated lung adenocarcinoma transcript 1-knockdown
cells compared with negative control cells. |
Table III.
Deregulated genes in
metastasis-associated lung adenocarcinoma transcript 1-knockdown
cells compared with negative control cells.
| Genbank access
number | Gene symbol | Microarray fold
change | qPCR fold change |
|---|
| NM_003881 | WISP2 | −17.73 | −10.38 |
| NM_024690 | MUC16 | 4.81 | 3.34 |
| NM_014795 | ZEB2 | −5.87 | −2.66 |
| NM_004530 | MMP2 | −5.17 | −3.43 |
| NM_005940 | MMP11 | −4.32 | −5.07 |
| NM_001025370 | VEGFA | −7.93 | −4.92 |
| NM_002632 | PGF | −4.87 | −8.38 |
| NR_002196 | H19 | −4.47 | −3.35 |
| NM_005235 | ERBB4 | −20.57 | −12.87 |
| NM_003068 | SNAI2 |
9.01 |
3.40 |
| NM_024680 | E2F8 |
4.63 |
7.98 |
| NM_001798 | CDK2 |
5.67 |
8.54 |
| NM_001255 | CDC20 |
4.52 |
5.67 |
| NM_000679 | ADRA1B |
4.26 |
7.82 |
| NM_000800 | FGF1 |
4.68 |
5.14 |
| NM_138764 | BAX |
4.48 |
2.05 |
| NM_054034 | FN1 | −23.30 | −3.79 |
| NM_022045 | MTBP |
4.70 |
5.55 |
| NM_018098 | ECT2 |
4.29 |
3.87 |
Discussion
MALAT1 was one of the first cancer-associated
lncRNAs to be identified, and is widely expressed in normal human
tissues. In 2003, Ji et al reported for the first time that
MALAT1 was upregulated in non-small cell lung cancer and was a
prognostic parameter for patient survival (8). Subsequent studies showed that MALAT1 was
also associated with the formation and progression of several other
types of tumors (9–12,14–18).
However, the role of MALAT1 in ovarian cancer remains unknown.
The present study first investigated the effect of
MALAT1 inhibition on SKOV3 cells, and the in vitro and in
vivo data suggested that MALAT1 inhibition suppressed
tumorigenicity in SKOV3 cells. A limitation to the present study
was the lack of clinical samples available. Additional studies with
clinical samples are warranted.
The gene expression in KD cells was then compared
with control cells, and a large number of genes that were altered
in association with decreased MALAT1 expression were identified.
The results of the RT-qPCR validation showed that MALAT1 inhibition
altered the expression levels of genes that have been reported to
be associated with cell proliferation (placental growth factor,
cyclin-dependent kinase 2, cell division cycle 20, adrenoceptor α
1B and fibroblast growth factor 1), metastasis [WNT1 inducible
signaling pathway protein 2, mucin 16, cell surface associated,
zinc finger E-box binding homeobox 2 (ZEB2), matrix
metallopeptidase 2, matrix metallopeptidase 11, vascular
endothelial growth factor A, H19, imprinted maternally expressed
transcript (non-protein coding), snail family zinc finger 2 (SNAI2)
and fibronectin 1], and apoptosis [erb-b2 receptor tyrosine kinase
4 and Bcl-2-associated X protein (BAX)]. Among these genes, ZEB2,
SNAI2 and BAX have been reported to be possible targets of MALAT1
(10,11). The mechanism of regulation of gene
expression by MALAT1 remains unclear. Tripathi et al
reported that MALAT1 regulated alternative splicing by modulating
SR splicing factor phosphorylation (19). However, this phenomenon was not
observed in lung cancer cells (20).
Additional studies are required in order to clarify the mechanisms
underlying MALAT1.
In conclusion, MALAT1 inhibition significantly
suppressed tumorigenity in ovarian cancer cells and the inhibition
of MALAT1 altered the expression of a number of genes associated
with cell proliferation, metastasis and apoptosis. Overall, the
findings of the present study define a major oncogenic role for
MALAT1, which may provide an attractive novel target for
therapeutic intervention in ovarian cancer.
Acknowledgements
The present study was supported by the National High
Technology Research and Development Program of China, Ministry of
Science and Technology of the People's Republic of China, Beijing,
China (grant no., 2012AA02A507).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holschneider CH and Berek JS: Ovarian
cancer: Epidemiology, biology, and prognostic factors. Semin Surg
Oncol. 19:3–10. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mørch LS, Løkkegaard E, Andreasen AH,
Krüger-Kjaer S and Lidegaard O: Hormone therapy and ovarian cancer.
JAMA. 302:298–305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin HW, Tu YY, Lin SY, Su WJ, Lin WL, Lin
WZ, Wu SC and Lai YL: Risk of ovarian cancer in women with pelvic
inflammatory disease: a population-based study. Lancet Oncol.
12:900–904. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ogawa Y, Sun BK and Lee JT: Intersection
of the RNA interference and X-inactivation pathways. Science.
320:1336–1341. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo F, Li Y, Liu Y, Wang J, Li Y and Li G:
Inhibition of metastasis-associated lung adenocarcinoma transcript
1 in CaSki human cervical cancer cells suppresses cell
proliferation and invasion. Acta Biochim Biophys Sin (Shanghai).
42:224–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY,
Chen SY and Li ZG: MALAT1 promotes colorectal cancer cell
proliferation/migration/invasion via PRKA kinase anchor protein 9.
Biochim Biophys Acta. 1852:166–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cho SF, Chang YC, Chang CS, Lin SF, Liu
YC, Hsiao HH, Chang JG and Liu TC: MALAT1 long non-coding RNA is
overexpressed in multiple myeloma and may serve as a marker to
predict disease progression. BMC Cancer. 14:8092014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Su L, Chen X, Li P, Cai Q, Yu B,
Liu B, Wu W and Zhu Z: MALAT1 promotes cell proliferation in
gastric cancer by recruiting SF2/ASF. Biomed Pharmacother.
68:557–564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
















