Introduction
Breast cancer is the most prevalent cause of
mortality in women worldwide, with risk factors that include age,
mutation of breast cancer (BRCA)1/BRCA2 genes, age at first full
term pregnancy and use of estrogen or progesterone (1). More than 1,300,000 women worldwide are
diagnosed with breast cancer each year, and 450,000 women succumb
to the disease (2). Standard
screening methods include mammography and physical examination
(3); however, it is understood that
up to 40% of early breast cancer cases go undetected using these
methods (4). Therefore, histological
evaluation is currently the gold standard for the detection of
breast cancer (5).
Breast cancer is a genetically and clinically
heterogeneous disease (6). The
histological and clinical outcomes of extensive research indicate
that the properties of breast tumors differ significantly between
individuals. This has resulted in a novel form of tumor
classification that depends on molecular markers (6). Numerous types of molecular markers based
on proteins and nucleotide sequences have been studied and
developed to aid in determining the diagnosis and prognosis of
breast cancer (7). However, only 3 or
4 are currently used in practice, including the estrogen receptor,
progesterone receptor and human epidermal growth factor receptor 2
(8). Thus, it is evident that
additional makers are required.
Glycosylation involves the formation of complex and
heterogeneous structures by stepwise attachment of oligosaccharides
and is the most common post-translational modification of proteins
and lipids (9). Previous studies have
indicated that carcinogenesis is associated with alterations in
glycosylation patterns (10,11). The variety of glycan structures and
their changes during carcinogenesis have great potential for
screening and diagnosis (12).
However, to date, the majority studies have focused on identifying
protein and nucleotide sequence markers (13).
To analyze glycosylation in clinical biopsy
specimens, the majority of previous studies have used mass
spectrometry (MS) and high-performance liquid chromatography (HPLC)
systems rather than a simple format permitting routine screening
(14). The former strategies require
specialized techniques and equipment, in addition to lengthy sample
preparation. In the present study, alterations in glycosylation
patterns were investigated as a function of stage of tumor
development in breast tissue using an enzyme-linked lectin assay
(ELLA) system. Lectins are carbohydrate-binding proteins that
recognize specific glycan structures (14). ELLA is the simplest method to analyze
lectins; however lectin blotting and lectin microarrays may also be
used (15). To date, ELLA systems
have been used to investigate a limited number of materials, with
purified glycoproteins analyzed most commonly (16,17). There
are few previous studies that have analyzed glycan structures in
clinical tissue biopsy specimens (18). To the best of our knowledge, the
present study demonstrates for the first time that the ELLA system
is capable of providing clear evidence of tumor development in
tissue biopsy specimens.
Materials and methods
Clinical samples
A total of 11 breast tissue sets, consisting of
paired cancer (CC) and normal (CN) tissues, were obtained from
patients who underwent surgical resection at the Ewha Womans
University Mokdong Hospital (Yangcheon-Gu, South Korea). The cancer
stage, age, and anti-hormone-, chemo- and radiotherapy histories of
each patient are presented in Table
I. Chemotherapy (adriamycin + cyclophosphamide) and hormone
therapy (Tamoxen, Tamoxen + Zoladex or Lenara) were conducted
following tissue resection, and the stage of each cancer was
determined by the tumor-node-metastasis staging system (19). All specimens were immediately frozen
and stored at −80°C for subsequent analysis. The present study was
conducted with the approval of the Ewha Womans University Mokdong
Hospital Institutional Review Board (approval no., EUMC
2014-11-013). Patient samples were obtained with written informed
consent from the patients.
 | Table I.Characteristics of analyzed
specimens. |
Table I.
Characteristics of analyzed
specimens.
|
|
|
| Therapy received |
|---|
|
|
|
|
|
|---|
| Specimen no. | Cancer stage | Patient age,
years | Anti-hormone | Chemotherapy | Radiotherapy |
|---|
| Stage 0-I |
|
|
|
|
|
| 1 | 0 | 44 | O |
|
|
| 2 | 0 | 42 | O |
| O |
| 3 | IA | 62 | O |
| O |
| 4 | IA | 50 |
| O |
|
| Stage II–III |
|
|
|
|
|
| 5 | IIA | 47 |
| O |
|
| 6 | IIA | 70 |
| O |
|
| 7 | IIA | 77 | O |
|
|
| 8 | IIA | 31 |
| O |
|
| 9 | IIB | 39 |
| O |
|
| 10 | IIB | 47 |
| O |
|
| 11 | IIIA | 44 |
| O |
|
Preparation of tissue lysates from
biopsies
Tissue lysates were prepared as previously described
(18), with modification. Biopsy
tissues were cut with scissors and mixed with lysis buffer [100 mM
Tris-Cl (pH 7.4; Duchefa Biochemie BV, Haarlem, Netherlands); 150
mM NaCl (Duchefa Biochemie BV); 1 mM ethylenediaminetetraacetic
acid-Na (Duchefa Biochemie BV); and 1% Triton X-100
(USB®; affymetrix, Santa Clara, CA, USA)]. The tissues
were then disrupted with a Dounce homogenizer (Duran Group GmbH,
Mainz, Germany) and centrifuged at 12,000 × g for 10 min to remove
debris and precipitated substances. The supernatants were
subsequently collected. Protein concentrations were determined
using a Pierce bicinchoninic acid protein assay kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), with bovine serum albumin
serving as a control.
ELLA
Wells of a 96-well microplate (Greiner Bio-One,
Frickenhausen, Germany) were coated with 1,600, 800, 400, 200 or
100 ng of tissue lysate protein per well at 4°C, and blocked with
5% oxidized bovine serum albumin (BSA; GenDEPOT, Katy, TX, USA) in
Tris-buffered saline containing 0.1% Tween 20 (TBS-T; Duchefa
Biochemie BV) for 4 h at room temperature. The cell lysates were
mixed with carbonate buffer (pH 9.4) and then incubated for 20 h at
4°C to coat the 96-well plate.. BSA treated with sodium
meta-periodate (Sigma-Aldrich, St. Louis, MO, USA) was prepared as
previously described (17).
Subsequently, the wells were incubated with biotinylated
concanavalin A (Con A; catalog no., B-1005), Ricinus
communis Agglutinin I (RCA I; catalog no., B-1085), Aleuria
aurantia lectin (AAL; catalog no., B-1395) or Maackia amurensis
lectin II (MAL II; catalog no., B-1265) (Vector Laboratories, Inc.,
Burlingame, CA, USA) for 80 min at 37°C, followed by
poly-horseradish peroxidase (HRP)-conjugated streptavidin (catalog
no., N200; Pierce™; Thermo Fisher Scientific, Inc.) for 40 min at
37°C. All lectins were prepared in a reaction buffer (TBS-T
containing 0.5% oxidized BSA) at a concentration of 1 µg/ml lectin.
HRP-conjugated streptavidin was diluted 1:5,000 with 0.5% oxidized
BSA in TBS-T. The reactions were developed with o-phenylenediamine
(Sigma-Aldrich) and optical density (OD) values were measured at
492 nm using a 96-well plate reader (SUNRISE; Tecan Group Ltd.,
Männedorf, Switzerland).
Con A affinity chromatography
Con A affinity chromatography was performed as
previously described (20), with
modification. A 0.8×4.0 cm Poly-Prep chromatography column
(Bio-Rad, Hercules, CA, USA) packed with 0.5 ml Con A Sepharose 4B
resin (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) was
equilibrated with binding buffer [20 mM Tris (pH 7.4; Duchefa
Biochemie BV) containing 0.5 M NaCl (Duchefa Biochemie BV), 1 mM
MgCl2 (Sigma-Aldrich) and 1 mM CaCl2
(Sigma-Aldrich)]. Tissue lysates were diluted with binding buffer
to 20 µg/ml of protein and loaded into the column. Thereafter, the
column was washed with 5 resin-bed volumes of binding buffer.
Proteins bound to the Con A resin were eluted with 5 resin-bed
volumes of elution buffer [20 mM Tris (pH 7.4) containing 0.5 M
NaCl and 0.5 M methyl-α-D-glucopyranoside (Sigma-Aldrich)]. The
elution fractions were subsequently examined using the ELLA
system.
Lectin blotting
Lectin blots were performed as previously described
(18). Samples containing 1,000 ng
protein per well were loaded and fractionated on a 12% acrylamide
gel. The fractionated proteins were transferred to a polyvinylidene
difluoride membrane (EMD Millipore, Billerica, MA, USA), and
blocked with 5% oxidized BSA in TBS-T for 4 h. The mannosylated
proteins were detected with Con A (1 µg/ml, prepared in 0.5%
oxidized BSA in TBS-T), followed by HRP-conjugated streptavidin
(diluted to 1:5,000 with 0.5% oxidized BSA). The membranes were
washed three times with TBS-T between reactions, and all reactions
were performed at room temperature. The membrane was developed
using an enhanced chemiluminescence kit (AbClon, Inc., Seoul, South
Korea), and the bands of mannosylated proteins were visualized on
X-ray film (Kodak, Rochester, NY, USA).
Statistical analysis
The statistical significance of differences between
groups was determined by two-tailed Student's t-tests using
GraphPad Prism version 5.01 software (GraphPad Software, Inc., La
Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference. Specificity was calculated as
follows: Specificity = number of true negatives × 100 / number of
true negatives + number of false positives. Sensitivity was
calculated by the following: Sensitivity = number of true positives
× 100 / number of true positives + number of false negatives. To
determine specificity and sensitivity, the cut-off value was set at
the highest value of CN at each cancer stage, and values higher
than the cut-off values were considered as positive for cancer.
Results
Subgrouping according to cancer
stage
Pairs of CN and CC tissues were obtained from each
patient with breast cancer. Lysates were prepared from 11 pairs of
specimens (Table I) and were used to
compare glycosylation levels in the CNs and CCs. The specimens were
divided into stage 0-I and stage II–III, and the glycosylation
levels of CNs and CCs in each specimen group were measured by ELLA.
Stage 0-I tumors were those not yet metastasized regionally and
<20 mm in size, while stage II–III tumors were those tending to
have spread to axillary lymph nodes or ranging in size from 20–50
mm.
Optimal protein coating for ELLAs
Con A, RCA I, MAL II and AAL recognize mannosylation
(preferentially binding to the high-mannose-type Manα1-6Man or
Manα1-3Man), galactosylation (preferentially binding to
Galβ1-4GlcNAc), sialylation (preferentially binding to
Siaα2-3Galβ1-4GlcNAc) and fucosylation (preferentially binding to
Fucα1-6GlcNAc or Fucα1-3GlcNAc), respectively (21,22). To
compare the OD values of the CNs and CCs as a function of the
amount of protein used for coating, a 96-well plate was coated with
1,600, 800, 400, 200 or 100 ng of protein per well. A total of 4
pairs of CN and CC (specimens 6, 7, 9 and 10, as presented in
Table I) were used for the ELLAs. As
shown in Fig. 1, no significant
differences were observed between the OD values of CN and CC when
wells were coated with 1,600 ng of protein (P>0.05; Fig. 1). However, CCs yielded significantly
higher values than CNs overall when certain amounts ≤800 ng of
protein per well were used for coating (P<0.05; Fig. 1). When MAL II was used for the
detection of sialylation, the difference between CNs and CCs
increased with a decreasing amount of protein coating. For the
subsequent ELLAs, 100 ng of protein per well was selected (Fig. 2).
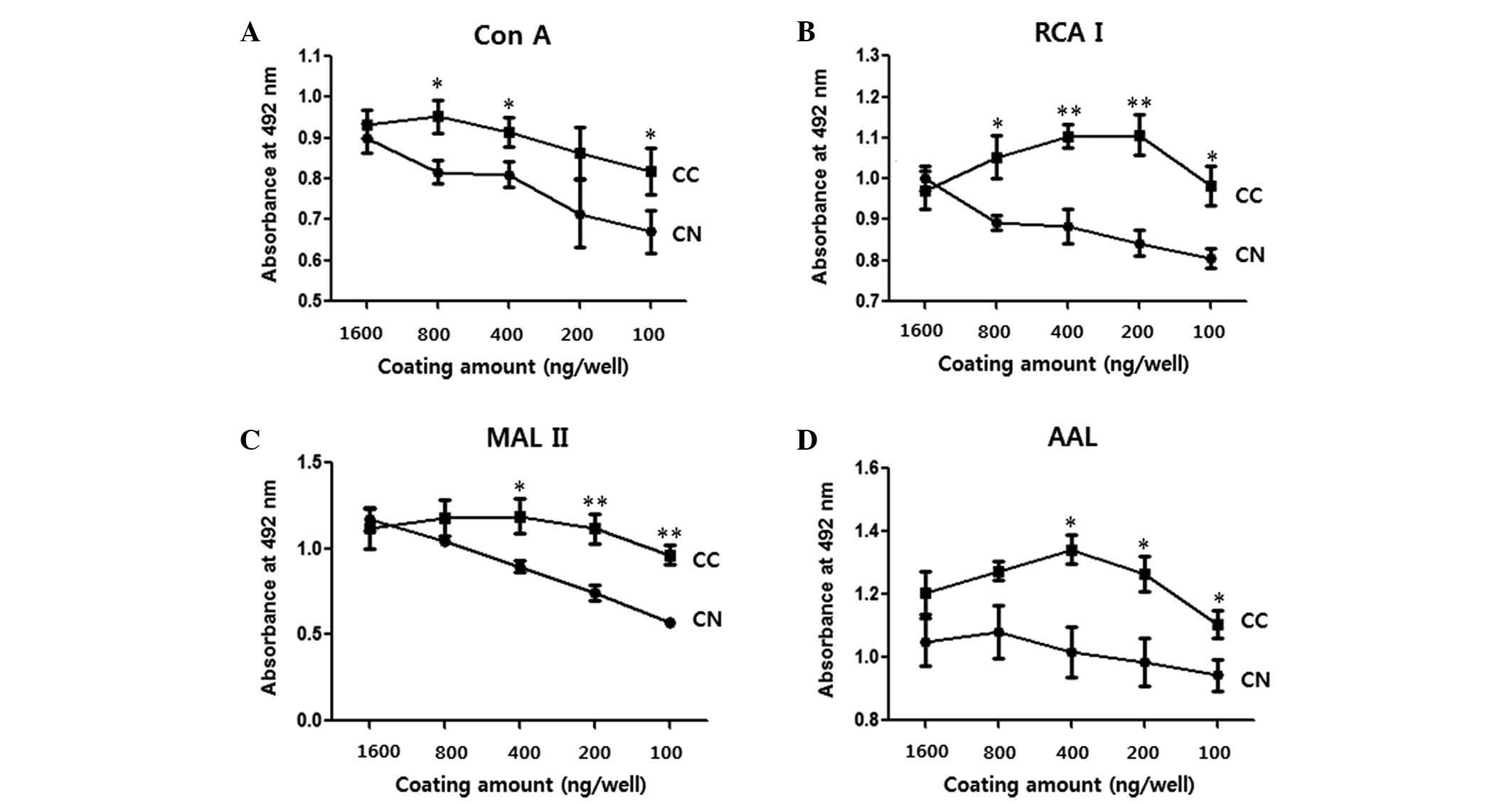 | Figure 1.Difference in OD values between CNs
and CCs as a function of the amount of protein coating. Each well
of a 96-well plate was coated with 1,600, 800, 400, 200 or 100 ng
of cell lysate protein, and enzyme-linked lectin assays were
performed. (A) Con A, (B) RCA I, (C) MAL II and (D) AAL were used
to detect mannosylation, galactosylation, sialylation and
fucosylation, respectively. Data are presented as the mean ±
standard error. *P<0.05 and **P<0.01. OD, optical density;
CN, normal tissue; CC, cancer tissue; Con A, concanavalin A; RCA I,
Ricinus communis Agglutinin I; MAL II, Maackia
amurensis lectin II; AAL, Aleuria aurantia lectin. |
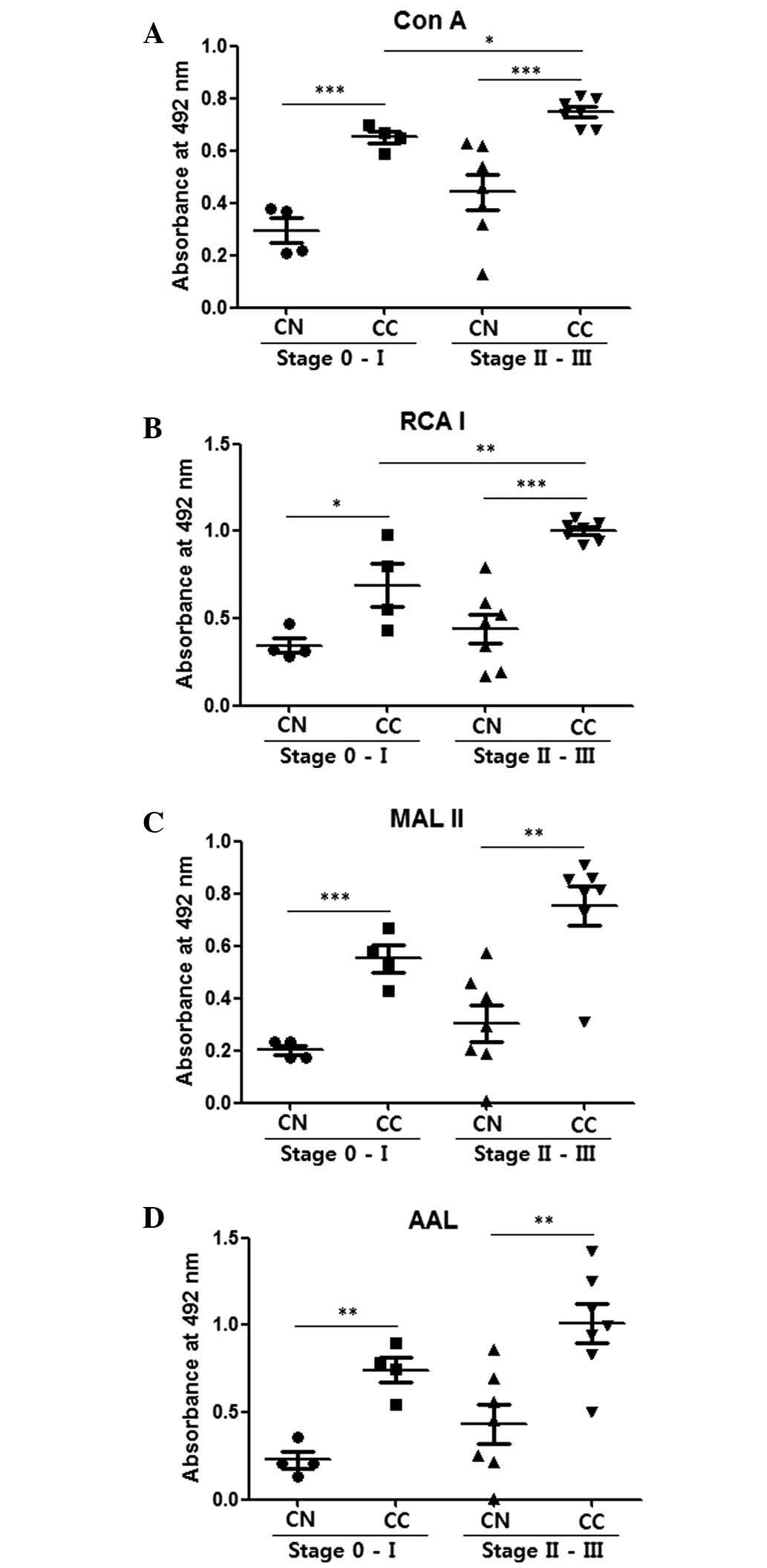 | Figure 2.Comparison of CNs and CCs by
enzyme-linked lectin assays with 100 ng cell lysate protein
coating. (A) Con A, (B) RCA I, (C) MAL II and (D) AAL were used to
detect mannosylation, galactosylation, sialylation and
fucosylation, respectively. The central line represents the median,
and the top and bottom lines represent the 75th and 25th
percentiles, respectively. *P<0.05, **P<0.01 and
***P<0.001. CN (stage 0-I), n=4; CC (stage 0-I), n=4; CN (stage
II–III), n=7; CC (stage II–III), n=7. CC, cancer tissue; CN, normal
tissue; Con A, concanavalin A; RCA I, Ricinus communis
Agglutinin I; MAL II, Maackia amurensis lectin II; AAL,
Aleuria aurantia lectin. |
Glycosylation levels of CNs and
CCs
Fig. 2 compares
glycosylation levels in CNs and CCs in the stage 0-I and stage
II–III groups. The CCs demonstrated significantly higher values for
mannosylation, galactosylation, sialylation and fucosylation
compared with the CNs for the stage 0-I and stage II–III groups
(P<0.05; Fig. 2). Stage II–III CCs
also demonstrated significantly higher mannosylation (P=0.015)
(Fig. 2A) and galactosylation
(P=0.009; Fig. 2B) levels compared
with the stage 0-I CCs. These results indicate that ELLAs have the
ability to distinguish stage II–III breast cancer from stage
0-I.
Similar differences were also observed between stage
II–III CNs and CCs with fractions from Con A affinity columns
enriched for glycoproteins with mannose-type glycans (Fig. 3). These results indicate that CCs have
markedly higher levels of glycosylation compared with the
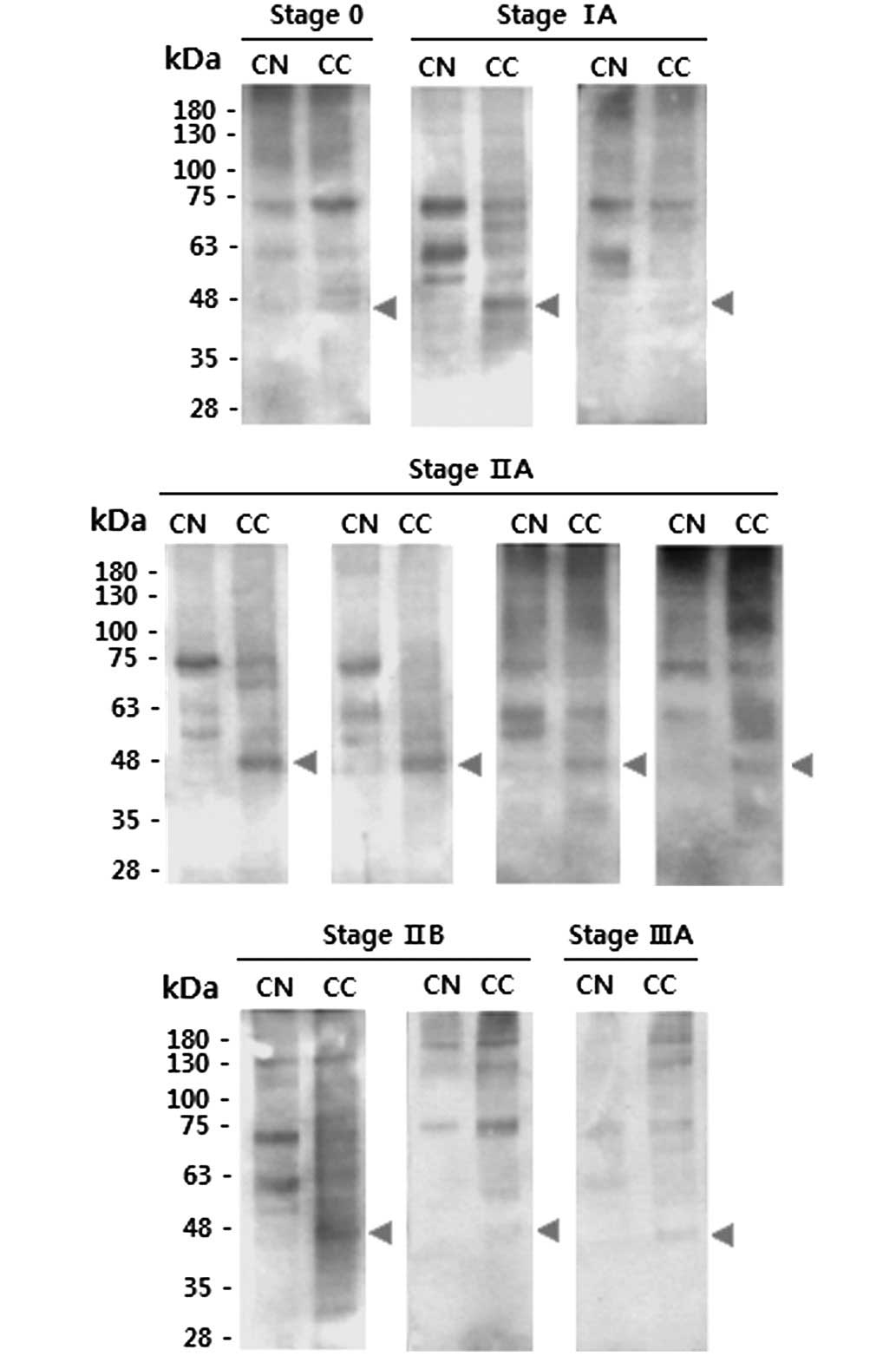
equivalent CNs. Banding patterns of CCs were compared with those of
the corresponding CNs following lectin blotting. Overall, the
protein bands detected by Con A were stronger in the CCs compared
with the CNs (Fig. 4). The blotting
results indicated that the CCs generally contained lectin-binding
proteins with a mass of ~48 kDa, whilst such proteins were absent
from the CNs.
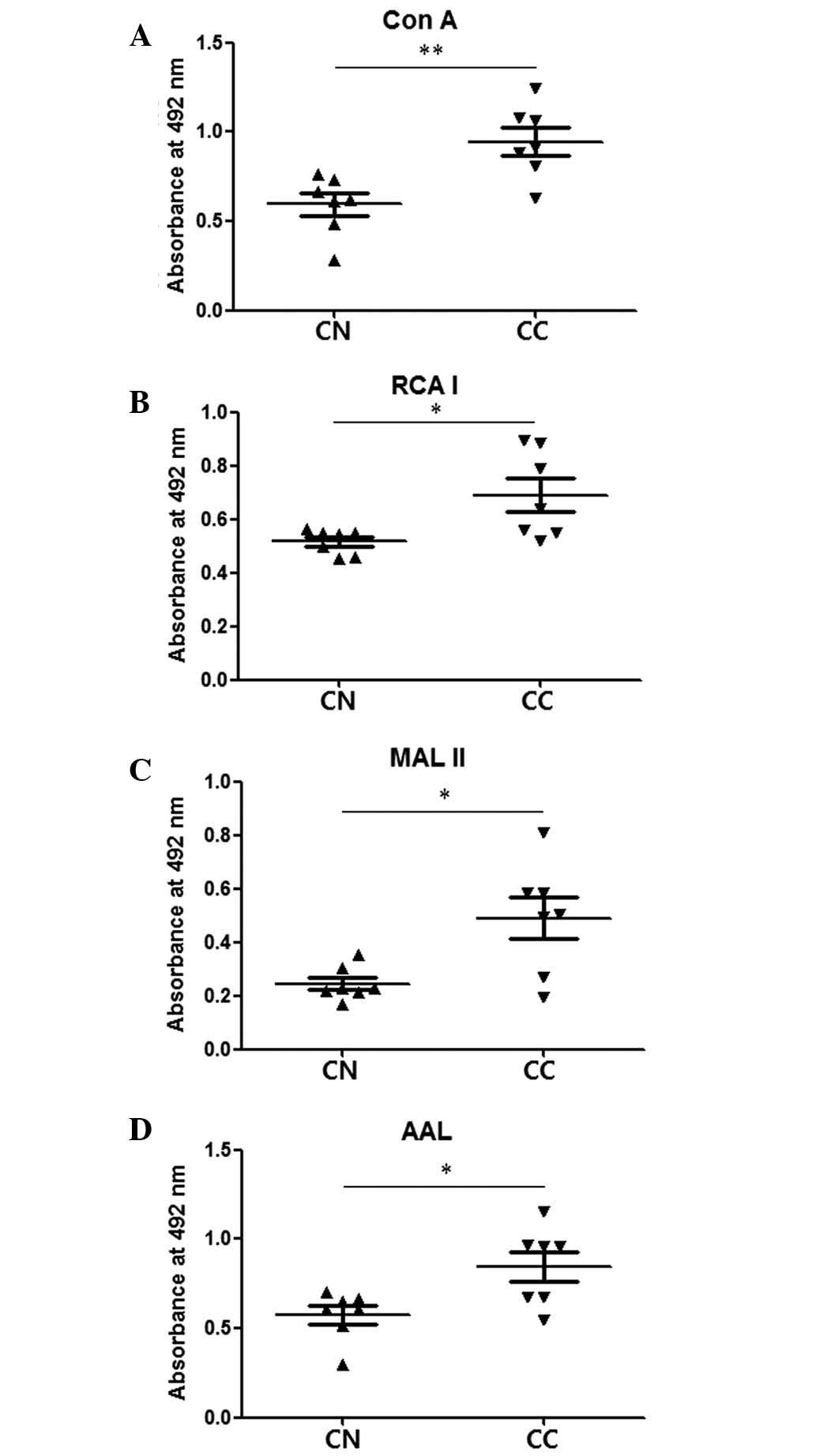 | Figure 3.Comparison of elution fractions of CNs
and CCs obtained by Con A affinity chromatography. Lysates of stage
II–III CNs and CCs were purified by Con A affinity chromatography,
and elution fractions were examined by enzyme-linked lectin assay.
(A) Con A, (B) RCA I, (C) MAL II and (D) AAL were used to detect
mannosylation, galactosylation, sialylation and fucosylation,
respectively. The central line represents the median, and the top
and bottom lines represent the 75th and 25th percentiles,
respectively. *P<0.05. CN (stage II–III), n=7; CC (stage
II–III), n=7. CC, cancer tissue; CN, normal tissue; Con A,
concanavalin A; RCA I, Ricinus communis Agglutinin I; MAL
II, Maackia amurensis lectin II; AAL, Aleuria
aurantia lectin. |
Sensitivity and specificity of
ELLAs
The sensitivity of each ELLA for discriminating
between CCs and CNs when specificity was set at 100% was
investigated (Table II). To set 100%
specificity, the highest OD value of the CNs was used as a cut-off
value, and thus the sensitivity of each lectin was evaluated in
conditions where there were no false-positives among the CNs. As
presented in Table I, Con A had 100%
sensitivity for glycans in stage 0-I and stage II–III. The other
lectins had sensitivities ranging from 71–100%. In addition, it was
confirmed that all CCs had higher values than the corresponding
CNs, with the exception of one case (specimen 2, RCA I, Table III). These results demonstrate that
ELLAs were able to detect CCs with high specificity and
sensitivity.
 | Table II.Sensitivity and specificity of
enzyme-linked lectin assays. |
Table II.
Sensitivity and specificity of
enzyme-linked lectin assays.
| Parameter | Con A, % | RCA I, % | MAL II, % | AAL, % |
|---|
| Stage 0-I |
|
|
|
|
|
Sensitivity | 100 | 75 | 100 | 100 |
|
Specificity | 100 | 100 | 100 | 100 |
| Stage II–III |
|
|
|
|
|
Sensitivity | 100 | 100 | 86 | 71 |
|
Specificity | 100 | 100 | 100 | 100 |
 | Table III.Comparison of OD values of CN and CC
in individual specimen. |
Table III.
Comparison of OD values of CN and CC
in individual specimen.
|
| Con A | RCA I | MAL II | AAL |
|---|
|
|
|
|
|
|
|---|
| Specimen no. | CN | CC | ↑/↓ | CN | CC | ↑/↓ | CN | CC | ↑/↓ | CN | CC | ↑/↓ |
|---|
| Stage 0-I |
|
| 1 | 0.37 | 0.67 | ↑ | 0.31 | 0.98 | ↑ | 0.17 | 0.58 | ↑ | 0.14 | 0.78 | ↑ |
| 2 | 0.38 | 0.65 | ↑ | 0.47 | 0.43 | ↓ | 0.23 | 0.43 | ↑ | 0.36 | 0.55 | ↑ |
| 3 | 0.21 | 0.70 | ↑ | 0.32 | 0.80 | ↑ | 0.18 | 0.67 | ↑ | 0.21 | 0.75 | ↑ |
| 4 | 0.22 | 0.59 | ↑ | 0.28 | 0.55 | ↑ | 0.23 | 0.53 | ↑ | 0.21 | 0.90 | ↑ |
| Stage II–III |
|
| 5 | 0.54 | 0.78 | ↑ | 0.48 | 0.92 | ↑ | 0.29 | 0.31 | ↑ | 0.22 | 0.50 | ↑ |
| 6 | 0.39 | 0.81 | ↑ | 0.17 | 1.05 | ↑ | 0.08 | 0.86 | ↑ | 0.05 | 0.94 | ↑ |
| 7 | 0.62 | 0.80 | ↑ | 0.79 | 0.98 | ↑ | 0.58 | 0.81 | ↑ | 0.86 | 0.96 | ↑ |
| 8 | 0.63 | 0.68 | ↑ | 0.59 | 1.08 | ↑ | 0.46 | 0.80 | ↑ | 0.56 | 1.10 | ↑ |
| 9 | 0.32 | 0.74 | ↑ | 0.34 | 1.02 | ↑ | 0.20 | 0.73 | ↑ | 0.26 | 0.83 | ↑ |
| 10 | 0.46 | 0.68 | ↑ | 0.52 | 0.94 | ↑ | 0.40 | 0.91 | ↑ | 0.70 | 1.43 | ↑ |
| 11 | 0.13 | 0.75 | ↑ | 0.19 | 1.03 | ↑ | 0.19 | 0.86 | ↑ | 0.45 | 1.25 | ↑ |
Discussion
Glycan structures, including sialyl
LewisA, sialyl LewisX and
Thomsen-Friedenreich (TF) antigen are observed more frequently on
the cell membranes of breast cancer tissues compared with normal
tissues (23,24). The increased sialyl LewisA
and sialyl LewisX structures observed in breast cancer
are considered to be involved in lymph node metastasis (22,25).
Overexpression of sialyl LewisA, sialyl
LewisX and TF antigen has also been reported in other
types of cancer (26). Similarly,
increased terminal sialylation and core fucosylation levels have
been detected in several forms of cancer, including liver,
colorectal and ovarian cancer (23).
Therefore, increased glycosylation and the formation of
cancer-specific glycan structures are considered as specific
features of cancerous tissue (27,28).
Alterations of glycan structures on tumor cell
membranes are associated with a poor prognosis and tumor
invasiveness (23). Therefore, the
majority of studies have focused on histochemical detection of
modifications in glycosylation on cell membranes or have used MS
and HPLC for similar purposes (29–31).
However, these approaches are of limited use for routine and
high-throughput analysis. ELLAs have reaction formats similar to
those of enzyme-linked immunosorbent assay (ELISA), using 96-well
plates and stepwise reactions with target substances. Unlike
ELISAs, which are based on antibody reactions and are widely used,
defects in lectin reaction conditions have hindered the use of
ELLAs for diagnosis (15). Recently,
the use of oxidized BSA (16,18) and polymer polyvinyl alcohol (15) has substantially improved the
specificity and sensitivity of ELLAs.
To date, there have been only a few attempts to
develop ELLAs for routine use in cancer diagnosis via tissue
biopsies. In a previous study utilizing ELLA, it was demonstrated
that the levels of sialylation and fucosylation in cytoplasmic
fractions of cervical cancer tissues were lower than in those of
normal cervical tissue, whilst no difference was reported in
mannosylation levels (18). However,
in the present study, it was observed that levels of mannosylation,
galactosylation, fucosylation and sialylation in the CCs were all
markedly higher than in the CNs (Fig.
2).
In the current study, the difference between the CNs
and CCs was greatest when Con A was used for the ELLA (Fig. 2), and the ELLA system had remarkable
sensitivity (71–100%) despite specificity being set at a maximum
value (100%). When Con A was used, the ELLAs had 100% sensitivity
and 100% specificity in discriminating between the CCs and CNs of
stages 0-I and II–III. Furthermore, it was confirmed that the Con A
protein bands were stronger in the CCs than in the CNs, and the CCs
generally contained lectin-binding proteins with a mass of ~48 kDa;
by contrast, these proteins were absent from the CNs (Fig. 4). Therefore, it is likely that the
48-kDa proteins are specific markers of breast tumor tissue and may
serve as potential targets for the treatment of breast cancer.
When Con A was used for affinity chromatography to
enrich glycoproteins with the high-mannose form of glycan, the
elution fractions from the CCs were observed to have higher levels
of mannosylation, galactosylation, sialylation and fucosylation
compared with those from the CNs (Fig.
3). This suggests that the glycoproteins with the high-mannose
form of glycans in CC tissue also have elevated galactosylation,
sialylation and fucosylation levels. A previous study reported that
enhanced levels of the high-mannose-type glycans are present in
serum from a mouse model of breast cancer and from breast cancer
patients (32). Together, these
results indicate that the presence of elevated levels of the
high-mannose-type glycan is a marker of breast cancer.
As the primary screening system, mammography has
contributed to reducing the risk of breast cancer (33). However, histological examination
following biopsy, which may cause stress for patients due to its
invasive nature, must be performed if a mass is palpable in the
breast or an abnormality is observed during a mammogram. Nipple
aspiration, originally developed by Papanicolaou et al
(34), is an almost non-invasive
method for detecting breast cancer, which yields cells from the
breast duct lining. Despite the great potential of fine needle
nipple aspiration biopsy for breast cancer screening, a high error
rate and inability to distinguish invasive cancer from non-invasive
cancer have hindered its widespread use (35). Nevertheless, nipple aspiration
continues to be used worldwide, particularly in developing
countries, due to its low cost. The ELLA system used in the present
study demonstrated extraordinarily high specificity and sensitivity
with only 100 ng of protein required for screening. It is therefore
anticipated that ELLA may serve as a complementary system for
breast cancer screening, in conjunction with nipple aspiration
biopsy.
In conclusion, the current study demonstrated the
potential value of the ELLA system as a highly specific and
sensitive method for diagnostic screening. Further study of the
alterations in glycosylation in breast cancer may provide
additional options for reliable diagnosis and prognosis of breast
cancer.
Acknowledgements
This study was supported by a grant from the Korean
Health Technology Research and Development Project, Ministry of
Health & Welfare, Republic of Korea (no. HI12C0050; www.htdream.kr/).
References
|
1
|
Donepudi MS, Kondapalli K, Amos SJ and
Venkanteshan P: Breast cancer statistics and markers. J Cancer Res
Ther. 10:506–511. 2014.PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kolb TM, Lichy J and Newhouse JH:
Comparison of the performance of screening mammography, physical
examination and breast US and evaluation of factors that influence
them: An analysis of 27, 825 patient evaluations. Radiology.
225:165–175. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alexander H, Stegner AL, Wagner-Mann C, Du
Bois GC, Alexander S and Sauter ER: Proteomic analysis to identify
breast cancer biomarkers in nipple aspirate fluid. Clin Cancer Res.
10:7500–7510. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ahmed HG: Impact of implementing grading
fine needle aspiration cytology in diagnosis of breast cancer
amongst sudanese women. Oman Med J. 26:99–103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Polyak K: Heterogeneity in breast cancer.
J Clin Invest. 121:3786–3788. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esteva FJ and Hortobagyi GN: Prognostic
molecular markers in early breast cancer. Breast Cancer Res.
6:109–118. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malhotra GK, Zhao X, Band H and Band V:
Histological, molecular and functional subtypes of breast cancers.
Cancer Biol Ther. 10:955–960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grunewald S, Matthijs G and Jaeken J:
Congenital disorders of glycosylation: A review. Pediatr Res.
52:618–624. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li M, Song L and Qin X: Glycan changes:
Cancer metastasis and anti-cancer vaccines. J Biosci. 35:665–673.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reis CA, Osorio H, Silva L, Gomes C and
David L: Alterations in glycosylation as biomarkers for cancer
detection. J Clin Pathol. 63:322–329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peracaula R, Barrabés S, Sarrats A, Rudd
PM and de Llorens R: Altered glycosylation in tumours focused to
cancer diagnosis. Dis Markers. 25:207–218. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van de Vijver MJ: Molecular tests as
prognostic factors in breast cancer. Virchows Arch. 464:283–291.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirabayashi J: Concept, strategy and
realization of lectin-based glycan profiling. J Biochem.
144:139–147. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thompson R, Creavin A, O'Connell M,
O'Connor B and Clarke P: Optimization of the enzyme-linked lectin
assay for enhanced glycoprotein and glycoconjugate analysis. Anal
Biochem. 413:114–122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim HJ, Lee SJ and Kim HJ: Antibody-based
enzyme-linked lectin assay (ABELLA) for the sialylated recombinant
human erythropoietin present in culture supernatant. J Pharm Biomed
Anal. 48:716–721. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim HJ, Lee DH, Kim DK, Han GB and Kim HJ:
The glycosylation and in vivo stability of human
granulocyte-macrophage colony-stimulating factor produced in rice
cells. Biol Pharm Bull. 31:290–294. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim HJ, Kim SC, Ju W, Kim YH, Yin SY and
Kim HJ: Aberrant sialylation and fucosylation of intracellular
proteins in cervical tissue are critical markers of cervical
carcinogenesis. Oncol Rep. 31:1417–1422. 2014.PubMed/NCBI
|
|
19
|
Harris JR: Natural history and staging of
breast cancer. Diseases of the Breast. Harris JR, Lippman ME,
Morrow M and Osborne CK: (1st). Lippincott-Raven. (Baltimore, PA).
457–459. 1996.
|
|
20
|
Chen GY, Chen CY, Chang MDT, Matsuura Y
and Hu YC: Concanavalin A affinity chromatography for efficient
baculovirus purification. Biotechnol Prog. 25:1669–1677.
2009.PubMed/NCBI
|
|
21
|
Clark D and Mao L: Cancer biomarker
discovery: Lectin-based strategies targeting glycoproteins. Dis
Markers. 33:1–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hirabayashi J, Kuno A and Tateno H:
Lectin-based structural glycomics: A practical approach to complex
glycans. Electrophoresis. 32:1118–1128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Renkonen J, Paavonen T and Renkonen R:
Endothelial and epithelial expression of sialyl Lewis(x) and sialyl
Lewis(a) in lesions of breast carcinoma. Int J Cancer. 74:296–300.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Imai J, Ghazizadeh M, Naito Z and Asano G:
Immunohistochemical expression of T, Tn and sialyl-Tn antigens and
clinical outcome in human breast carcinoma. Anticancer Res. 21(2B):
1327–1334. 2001.PubMed/NCBI
|
|
25
|
Jeschke U, Mylonas I, Shabani N,
Kunert-Keil C, Schindlbeck C, Gerber B and Friese K: Expression of
sialyl lewis X, sialyl Lewis A, E-cadherin and cathepsin-D in human
breast cancer: Immunohistochemical analysis in mammary carcinoma in
situ, invasive carcinomas and their lymph node metastasis.
Anticancer Res. 25(3A): 1615–1622. 2005.PubMed/NCBI
|
|
26
|
Christiansen MN, Chik J, Lee L, Anugraham
M, Abrahams JL and Packer NH: Cell surface protein glycosylation in
cancer. Proteomics. 14:525–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Häuselmann I and Borsig L: Altered
tumor-cell glycosylation promotes metastasis. Front Oncol.
4:282014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miyoshi E, Moriwaki K, Terao N, Tan CC,
Terao M, Nakagawa T, Matsumoto H, Shinzaki S and Kamada Y:
Fucosylation is a promising target for cancer diagnosis and
therapy. Biomolecules. 2:34–45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Van Elssen CH, Frings PW, Bot FJ, Van de
Vijver KK, Huls MB, Meek B, Hupperets P, Germeraad WT and Bos GM:
Expression of aberrantly glycosylated Mucin-1 in ovarian cancer.
Histopathology. 57:597–606. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu H, Zhang N, Wan D, Cui M, Liu Z and
Liu S: Mass spectrometry-based analysis of glycoproteins and its
clinical applications in cancer biomarker discovery. Clin
Proteomics. 11:142014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Holst S, Stavenhagen K, Balog CI, Koeleman
CA, McDonnell LM, Mayboroda OA, Verhoeven A, Mesker WE, Tollenaar
RA, Deelder AM and Wuhrer M: Investigations on aberrant
glycosylation of glycosphingolipids in colorectal cancer tissues
using liquid chromatography and matrix-assisted laser desorption
time-of-flight mass spectrometry (MALDI-TOF-MS). Mol Cell
Proteomics. 12:3081–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
de Leoz ML, Young LJ, An HJ, Kronewitter
SR, Kim J, Miyamoto S, Borowsky AD, Chew HK and Lebrilla CB:
High-mannose glycans are elevated during breast cancer progression.
Mol Cell Proteomics. 10:M110.0027172011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weedon-Fekjaer H, Romundstad PR and Vatten
LJ: Modern mammography screening and breast cancer mortality:
Population study. BMJ. 348:g37012014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Papanicolaou GN, Holmquist DG, Bader GM
and Falk EA: Exioliative cytology of the human mammary gland and
its value in the diagnosis of cancer and other diseases of the
breast. Cancer. 11:377–409. 1958. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Harigopal M and Chhieng DC: Breast
cytology: Current issues and future directions. The Open Breast
Cancer Journal. 2:81–89. 2010.
|