Introduction
MicroRNAs (miRNAs or miRs) are small, endogenous,
non-coding RNAs that act as post-transcriptional regulators by
binding to the 3′-untranslated region (UTR) of their target
messenger (m)RNAs, resulting in the degradation of the mRNA or its
translational inhibition (1–5).
miRNAs are 18–25 nucleotides in length, and are
generated by cytoplasmic RNase III Dicer from 70–100
nucleotides-long endogenous hairpin pre-miRNA precursors (2–5). miRNAs
have been highly conserved during evolution (6). Presently, ~2,588 human mature miRNAs
have been deposited in the miRBase (release 21, http://www.miRBase.org/). Accumulating evidence
indicates that miRNAs influence a variety of cellular functions,
including proliferation, differentiation and apoptosis (3,4,7). Furthermore, an increasing number of
miRNAs have been implicated in a variety of diseases such as cancer
(3). According to their function and
expression pattern, miRNAs may act as oncogenes or tumor
suppressors in cancer development and progression (3,7).
miR-152 is one of the miRNAs that have attracted
great interest in recent years, since it is implicated in various
types of cancer (8,9). Thus, it is of considerable significance
to understand the regulation and function of miR-152 in human
cancer. In the present study, the current knowledge of the
functions of miR-152 in cancer is reviewed, with an emphasis on its
regulation, targets and tumor suppressor role in human cancer.
Biogenesis and evolution of miR-152
miR-152 was first identified in mouse colon by
tissue-specific cloning in 2002 (10). miR-152 is a member of the miR-148/152
family, which includes miR-148a, miR-148b and miR-152. Notably, in
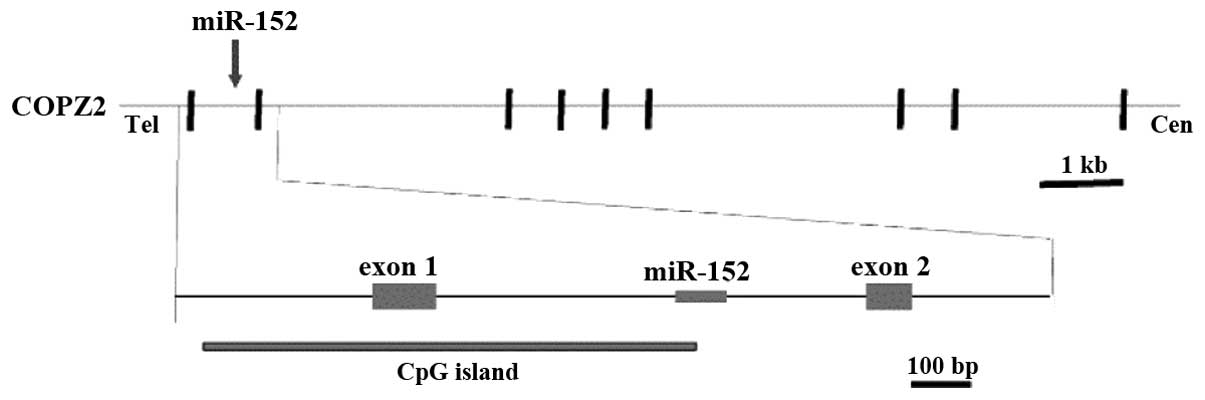
humans, the miR-152 gene is located on chromosome 17q21.32
(Fig. 1), within intron 1 of the
coatomer protein complex, subunit zeta 2 (COPZ2) gene, and a CpG
island is typically observed around its promoter region (8,9,11). Following transcription and cleavage by
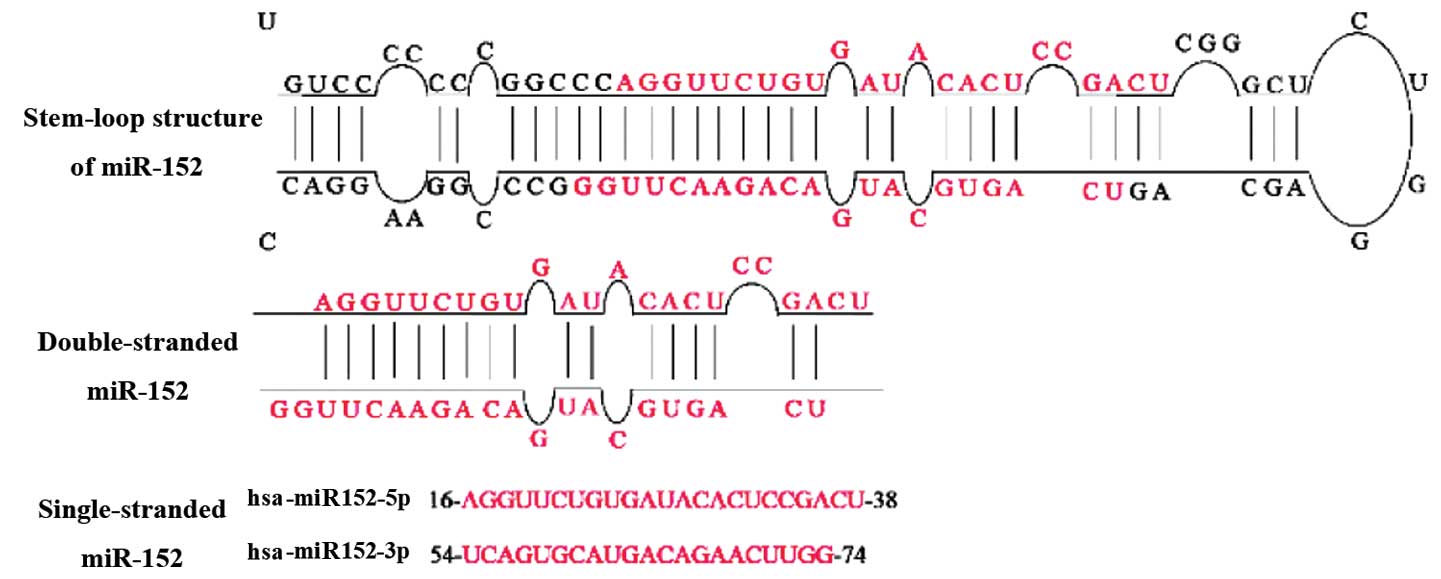
Drosha, pre-miR-152 is transported to the cytoplasm, where it is
further processed by Dicer to form a miR-152 duplex (5). Two different mature miR-152 sequences,
namely miR-152-5p and miR-152-3p, appear to be excised from
opposite arms of the miR-152 duplex (Fig.
2). miR-152-3p, which is excised from the 3′ arm of the hairpin
precursor, has been detected in more species than miR-152-5p.
The expression of miR-152 has been demonstrated in
several species, according to miRBase (http://www.mirbase.org/). It is noteworthy that the
mature miR-152 exhibits identical sequence in different species
(with the exception of the extension at its 3′ end), and also
shares an identical seed sequence across various species (Fig. 3A), which suggests that miR-152 is
important in certain gene regulatory networks. To assess the degree
to which miR-152 is conserved across species, a neighbor-joining
(NJ) tree was constructed in 10 representative animal species by
using genomic DNA sequences retrieved from miRBase (Fig. 3B). The miR-152 NJ tree clearly
revealed the existence of two main lineages, one of which contains
Homo sapiens, Canis familiaris, Ovis aries,
Sus scrofa, Mus musculus, Bos taurus,
Monodelphis domestica and Pan troglodytes, while the
other lineage comprises Fugu rubripes and Danio
rerio. These findings suggest that miR-152 is evolutionarily
conserved and the recent lineage-specific miR-152 may be common to
the old ancestral processor.
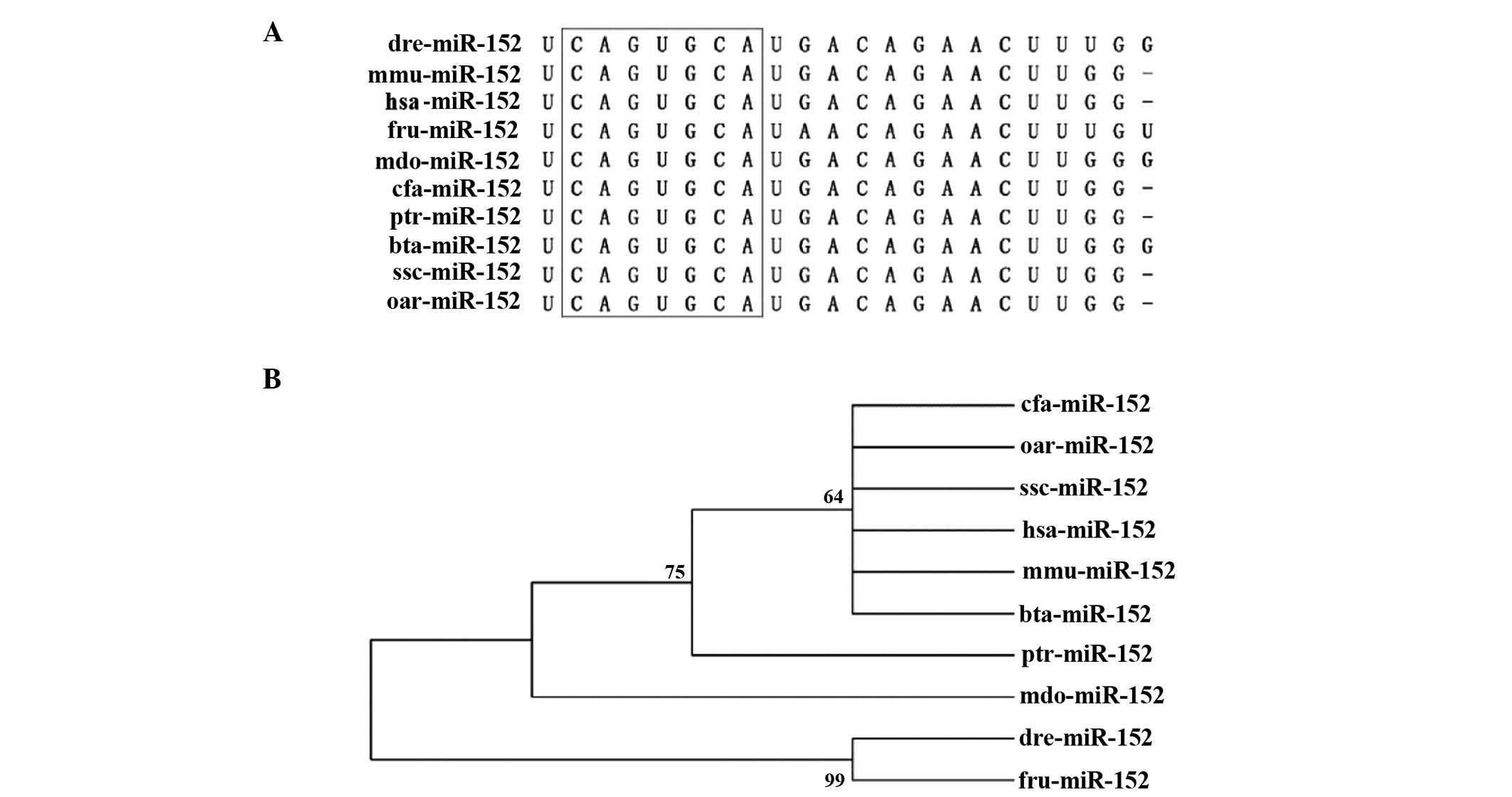 | Figure 3.Conservation of miR-152 across
species. (A) Sequence alignment of the mature miR-152 sequence in
10 different species. Seed sequences are highlighted in the black
box. (B) Phylogenetic tree of miR-152. Numbers on each branch refer
to the degree of reliability. miR, microRNA; hsa, Homo
sapiens; cfa, Canis familiaris; oar, Ovis aries;
ssc, Sus scrofa; mmu, Mus musculus; bta, Bos
taurus; mdo, Monodelphis domestica; ptr, Pan
troglodytes; fru, Fugu rubripes; dre, Danio
rerio. |
Experimentally validated targets of miR-152
in human cancer
The identification of miRNA targets and their
regulatory sequences is a complex problem in miRNA research, and a
considerable number of methods have been established to attempt
such an identification, which are classified into computational
(in silico) and experimental methods (12,13). The
prediction of miRNA targets using the current algorithms
implemented in computational methods always results in a large
number of false signals that do not reflect the situation in
vivo; therefore, the predicted miRNA targets must be validated
experimentally (12).
Hundreds of genes have been proposed as candidate
targets of miR-152 with high scoring when predicted by
computational programs such as PicTar (http://pictar.mde-berlin.de/) and TargetScan
(http://www.targetscan.org/). A number of
these genes have been further confirmed experimentally as targets
of miR-152 (Table I). Braconi et
al (14) first described that the
DNA methyltransferase 1 (DNMT1) gene is a direct target of miR-148a
and miR-152 by using luciferase reporter constructs, which revealed
that miR-152 could target the 3′-UTR of DNMT1, resulting in a
significant reduction of DNMT1 at both mRNA and protein levels.
This finding was further confirmed in subsequent studies on ovarian
cancer (15), endometrial cancer
(9), nickel sulphide (NiS)-induced
cell malignant transformation (16),
breast cancer (17), hepatitis B
virus-related hepatocellular carcinoma (18), pancreatic cancer (19) and prostate cancer (20). In addition, E2F transcription factor
3, mesenchymal to epithelial transition (MET),
rapamycin-insensitive companion of mechanistic target of rapamycin
(9), insulin-like growth factor 1
receptor (IGF-1R), insulin receptor substrate 1 (IRS1) (17), a disintegrin and metalloproteinase
metallopeptidase domain 17 (ADAM17) (21,22),
Kruppel-like factor 4 (KLF4) (23),
fibroblast growth factor 2 (FGF2) (24), wingless-related integration site
(Wnt1) (25), cluster of
differentiation (CD) 151 (26),
matrix metalloproteinase 3 (MMP3) (27) and transforming growth factor alpha
(28) have been identified as targets
of miR-152 in a wide array of human malignancies.
 | Table I.Experimental confirmed targets of
microRNA-152 in different types of cancer. |
Table I.
Experimental confirmed targets of
microRNA-152 in different types of cancer.
| Cancer | Expression | Biological
process | Target gene | Refs. |
|---|
| Endometrial
cancer | Downregulation | Inhibited cell
growth | DNMT1, E2F3, MET,
RICTOR | (9) |
|
Cholangiocarcinoma | Downregulation | Reduced cell
proliferation | DNMT1 | (14) |
| Ovarian cancer | Downregulation | Reduced cell
proliferation | DNMT1 | (15) |
| NiS-induced cell
malignant transformation | Downregulation | Inhibited cell
growth | DNMT1 | (16) |
| Breast cancer | Downregulation | Inhibited cell
proliferation, colony formation and tumor angiogenesis | DNMT1, IGF-1R,
IRS1 | (17) |
| Hepatocellular
carcinoma | Downregulation | Reduced cell
proliferation | DNMT1, Wnt1 | (18,25) |
| Pancreatic
cancer | Downregulation | Inhibited cell
proliferation | DNMT1 | (19) |
| Prostate
cancer | Downregulation | Decreased cell
growth, migration | DNMT1, TGFα | (20,28) |
| NSCLC | Downregulation | Reduced cell
proliferation, colony formation, migration and invasion | ADAM17, FGF2 | (22,24) |
| Glioblastoma | Downregulation | Reduced cell
proliferation, migration, invasion and pro-apoptosis | KLF4 | (23) |
| Gastric cancer | Downregulation | Inhibited cell
proliferation and motility | CD151 | (26) |
| Glioma | Downregulation | Reduced invasion
and invasion | MMP3 | (27) |
| Neuroblastoma | Upregulation | Increased
neuroblast differentiation and apoptosis | CHUK, CUL5,
GADD45A | (36) |
miR-152 as a tumor suppressor miRNA in
cancer
Particular miRNAs may act as oncogenes or tumor
suppressors depending on their expression pattern and function
(3,6).
Those miRNAs with increased expression in tumor cells may be
regarded as oncogenes, whereas downregulated miRNAs are considered
as tumor suppressors (29).
There has been a rapid increase in the number of
publications focusing on miR-152 in recent years, which revealed
that the expression of miR-152 was inhibited in a variety of
tumors, including ovarian (15),
endometrial (9) and breast cancer
(17). These findings suggest that
miR-152 may potentially function as a tumor suppressor in human
cancer. The main aim of the present review was to understand how
miR-152 interacts with its target genes and to identify the
potential role of miR-152 in cancer.
miR-152 mediates hypermethylation of
DNA and its CpG island in human cancer
Aberrant DNA hypermethylation of tumor suppressor
genes, global DNA hypermethylation (GDM) and disruption of histone
modification patterns are the three most important epigenetic
changes contributing to the malignant phenotype (30). In particular, DNA hypermethylation may
be important in the initiation of multiple types of cancer
(30,31). Huang et al (18) first reported that inhibition of
miR-152 could functionally result in GDM in hepatitis B
virus-related hepatocellular carcinoma cell lines. Using liquid
chromatography–mass spectrometry (MS)/MS, the authors identified
that the overexpression of miR-152 reduced GDM from 6.31 to 4.08%
in the HepG2 2.2.15 cell line, whereas miR-152 inhibitor increased
GDM from 4.55 to 5.88% in HepG2 cells. Underexpression of miR-152
also increased the DNA methylation level of the promoter region of
tumor suppressor genes such as glutathione S-transferase pi 1 and
cadherin 1 in these cells (18).
Azizi et al (19) demonstrated
that the overexpression of miR-152 decreased GDM to normal patterns
in pancreatic cancer cell lines and restored the expression of
tumor suppressor genes, including B-cell lymphoma 2/adenovirus E1B
19 kDa interacting protein 3 and secreted protein acidic and
cysteine rich, by 3.8 and 2.9-fold, respectively. These data
support a tumor suppressor role of miR-152 in the epigenetic
aberration observed in cancer.
An increasing number of publications indicate that
epigenetic silencing of tumor suppressor miRNAs by CpG island
hypermethylation is a common feature of different types of human
cancer (31,32). Hypermethylation of the CpG island of
miR-152 has been detected in 70 (97.1%) cases of primary
endometrial cancer (9). The
concordance between DNA hypermethylation around the CpG island and
underexpression of miR-152 was observed in 100% of the 70 cases of
primary endometrial cancer (9). These
results suggest that the hypermethylation of the CpG island of
miR-152 may downregulate its expression, and may be involved in
endometrial cancer. Due to the hypermethylation of its CpG island,
silencing of miR-152 expression and overexpression of DNMT1 were
also observed in NiS-transformed cells (16), breast cancer (17) and prostate cancer (20). Notably, there may be a crucial
functional crosstalk between miR-152 and DNMT1 via a
double-negative feedback regulatory loop, as speculated by Ji et
al (16) regarding the classic
‘chicken and egg’ argument. DNMT1 exerts a crucial role in setting
up and maintaining DNA methylation patterns in eukaryotic cells
(33). Once increased expression of
DNMT1 (‘egg’) occurs, DNMT1 is recruited to the miR-152 CpG island
promoter, where it increases DNA methylation, contributing to
reduced miR-152 expression (‘chicken’) (16). Furthermore, downregulated expression
of miR-152 further increases DNMT1 expression by reduced targeting
on DNMT1 3′-UTR (14–20). Therefore, epigenetic regulation of
miR-152/DNMT1 may be important in tumorigenesis. In mixed lineage
leukemia-rearranged infant acute lymphoblastic leukemia,
hypermethylation of the CpG island of miR-152 was reported to be
strongly correlated with a poor clinical outcome (34). Overall, hypermethylation of miR-152
may be considered as an epigenetic biomarker in human cancer.
miR-152 and its targets are associated
with cell proliferation in cancer
miRNAs with antiproliferative and pro-apoptotic
activity are likely to function as tumor suppressor genes (35). Antisense oligonucleotides targeting
miRNAs have been used to identify miRNA functions (36). In those studies, the inhibition of
miR-152 was observed to cause a decrease in cell growth in Hela
cells. In neuroblastoma samples, the expression of miR-152 was
upregulated, and miR-152 negatively controlled apoptosis by
downregulating pro-apoptotic genes such as conserved
helix-loop-helix ubiquitous kinase, cullin 5 and growth arrest and
DNA-damage-inducible, alpha (37). By
contrast, Zhou et al (9)
reported that cell proliferation was remarkably inhibited by
overexpression of miR-152 in ovarian cancer cells using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
Therefore, whether miR-152 acts as a tumor suppressor gene remains
controversial.
In previous studies, several miRNAs have been
demonstrated to affect target genes that are involved in the
control of cell proliferation and apoptosis (14–20). It
has been well established that phosphatidylinositol-3 kinase
(PI3K)/AKT and mitogen-activated protein kinase (MAPK)-mediated
signaling pathways are two of the most predominant signaling
pathways in human cancer, since they are involved in cell
proliferation, survival and metabolism (38,39). In
breast cancer, overexpression of miR-152 significantly inhibited
cell proliferation, colony formation and tumor angiogenesis by
targeting IGF-1R and IRS1, and suppressing their downstream AKT and
MAPK signaling pathways (17). KLF4
is a transcription factor that functions either as a tumor
suppressor gene or as an oncogene in different contexts, and is
critical for the control of essential cellular processes (40). In glioblastoma stem cells, miR-152
markedly inhibits cell proliferation, migration and invasion, and
promotes cell apoptosis by targeting KLF4 (23). In addition, miR-152 could inhibit the
expression of lectin, galactoside-binding, soluble, 3 by
downregulating KLF4, thus attenuating the activation of the MAPK
kinase 1/2 and PI3K signaling pathway (23). In non-small cell lung cancer (NSCLC),
Su et al (22) demonstrated
that restoration of miR-152 significantly reduced cell
proliferation, colony formation, migration and invasion partially
via targeting ADAM17 (also known as tumor necrosis factor-alpha
converting enzyme), which releases a variety of membrane-tethered
proteins, the majority of which are associated with pathological
processes such as cancer and inflammation (21). Another report revealed that the
ectopic overexpression of miR-152 markedly inhibited NSCLC cell
proliferation, promoted apoptosis, and suppressed migration and
invasion through targeting FGF2 (22). CD151, a transmembrane protein of the
tetraspanin family, participated in the mediation of tumor growth
and metastasis (41). miR-152 was
previously observed to be able to suppress the proliferation and
motility of gastric cancer cell lines by targeting CD151 (26). In addition, miR-152 is also able to
target Wnt1 and MMP3 (27) to inhibit
cell proliferation in liver cancer cells, thus reducing glioma cell
invasion and angiogenesis, respectively. Taking together, these
findings suggest that miR-152 may modulate a variety of cellular
processes such as cell proliferation, apoptosis and tumorigenesis
via the regulation of its target genes and the tumor suppressor
role of miR-152 in human cancer.
miR-152 in immune response
During the last 20 years, miRNAs have emerged as key
regulators of a wide range of biological processes, including cell
proliferation, differentiation, development and apoptosis (3,4,7). Recent studies indicate that specific
miRNAs are important in the immune system by modulating the
development of immune cells and regulating the expression of genes
that are critically involved in the immune response (42).
The innate immune system provides the first line of
defence against infections and natural killer (NK) cells are
critical mediators of the innate immune response (43). Human leucocyte antigen (HLA)-G is
important in the cellular immune response, since it inhibits NK
cell activity (44). miR-152 may
downregulate the expression of HLA-G by directly targeting its
3′-UTR, leading to increased NK cell-mediated cytosis (45). Dendritic cells (DCs) are professional
antigen-presenting cells, which bridge the innate and adaptive
immune responses (45).
Calcium/calmodulin-dependent protein kinase IIa (CaMKIIa), a major
downstream effector of calcium signaling, regulates the critical
stages of maturation and antigen-presentation capacity of human DCs
(46). miR-152 is capable of
inhibiting lipopolysaccharide-induced upregulation of major
histocompatibility complex II expression and DC-initiated
antigen-specific CD4+ T cell proliferation by targeting
CaMKIIa (47). These findings suggest
that miR-152 acts as a negative regulator in the immune system.
Therefore, in addition to human cancer, miR-152 is also important
in the innate immune response.
Therapeutic potential of miR-152 in
cancer
Aberrant miRNA expression is a common feature of
various types of human cancer, and miRNAs are crucial in the
development of cancer (3,7). As a result, numerous studies have
focused on miRNA-based therapeutics, some of which are undergoing
clinical trials in cancer patients (15). Targeting miRNAs may be used to control
the growth of cancer cells, and also to enhance the efficacy of
other therapies, such as reducing the drug resistance of tumors
(48).
Resistance of cancer cells to chemotherapeutics is a
clinical obstacle in the treatment of cancer patients (49). Cisplatin is the first-line
chemotherapy drug for multiple malignancies (50). Xiang et al (15) demonstrated that miR-152 was involved
in resistance to cisplatin in ovarian cancer. The authors confirmed
that overexpression of miR-152 increased cisplatin sensitivity of
SKOV3/DDP and A2780/DDP cells by inhibiting cell proliferation and
promoting cell apoptosis via direct suppression of DNMT1.
Therefore, miR-152 may serve as a therapeutic target for overcoming
cisplatin resistance in ovarian cancer. This application is also
likely to be used as a potential epigenetic therapeutic target in
other types of cancer.
Conclusion
miR-152 is well conserved in evolution and possesses
an identical seed sequence in different species. miR-152 may
repress multiple target genes, a number of which have been
validated by experimental methods. miR-152 binds to the 3′-UTR of
its target genes, which are associated with different signaling
pathways, thus leading to reduced cell proliferation and
pro-apoptosis. In addition, miR-152 is involved in tumorigenesis,
cell migration and invasion. miR-152 is located in the intron 1 of
the COPZ2 gene, and is surrounded by a CpG island. Hypermethylation
of the CpG island of miR-152 has been described in certain type of
human cancer, and it may account for the downregulation of miR-152.
These findings support the tumor suppressor role of miR-152 in
human cancer, and suggest that miR-152 may serve as a prognostic
biomarker and a therapeutic target in cancer patients.
However, the role of miR-152 in the progression of
human tumors remains to be fully understood, particularly the
mechanisms by which miR-152 contributes to tumorigenesis by binding
to different target genes in different types of cancer. Further
investigation on the function of miR-152 may lead to novel
diagnostic and therapeutic approaches for the treatment of human
cancer.
Acknowledgements
The present study was supported by Guangxi
University of Science and Technology (Liuzhou, China; grant no.
2014G020403). The authors would like to thank Professor Wei Tian
(Central South University, Changsha, Hunan, China) for his
assistance in proofreading the present manuscript.
References
|
1.
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
3.
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012.PubMed/NCBI
|
|
6.
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Zhou X, Zhao F, Wang ZN, Song YX, Chang H,
Chiang Y and Xu HM: Altered expression of miR-152 and miR-148a in
ovarian cancer is related to cell proliferation. Oncol Rep.
27:447–454. 2012.PubMed/NCBI
|
|
10.
|
Chen Y, Song YX and Wang ZN: The
microRNA-148/152 family: Multi-faceted players. Mol Cancer.
12:432013. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Tsuruta T, Kozaki K, Uesugi A, Furuta M,
Hirasawa A, Imoto I, Susumu N, Aoki D and Inazawa J: miR-152 is a
tumor suppressor microRNA that is silenced by DNA hypermethylation
in endometrial cancer. Cancer Res. 71:6450–6462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Ekimler S and Sahin K: Computational
methods for microRNA target prediction. Genes (Basel). 5:671–683.
2014.PubMed/NCBI
|
|
13.
|
Thomson DW, Bracken CP and Goodall GJ:
Experimental strategies for microRNA target identification. Nucleic
Acids Res. 39:6845–6853. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Braconi C, Huang N and Patel T:
MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor
suppressor gene expression by interleukin-6 in human malignant
cholangiocytes. Hepatology. 51:881–890. 2010.PubMed/NCBI
|
|
15.
|
Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu
G, Zhao R, Huang H, Wang X, Qiao Y, et al: miR-152 and miR-185
co-contribute to ovarian cancer cells cisplatin sensitivity by
targeting DNMT1 directly: A novel epigenetic therapy independent of
decitabine. Oncogene. 33:378–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Ji W, Yang L, Yuan J, Yang L, Zhang M, Qi
D, Duan X, Xuan A, Zhang W, Lu J, et al: MicroRNA-152 targets DNA
methyltransferase 1 in NiS-transformed cells via a feedback
mechanism. Carcinogenesis. 34:446–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y,
Qi YT, Xu Q, Li W, Lu B, et al: A regulatory circuit of
miR-148a/152 and DNMT1 in modulating cell transformation and tumor
angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 5:3–13.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Huang J, Wang Y, Guo Y and Sun S:
Down-regulated microRNA-152 induces aberrant DNA methylation in
hepatitis B virus-related hepatocellular carcinoma by targeting DNA
methyltransferase 1. Hepatology. 52:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Azizi M, Teimoori-Toolabi L, Arzanani MK,
Azadmanesh K, Fard-Esfahani P and Zeinali S: MicroRNA-148b and
microRNA-152 reactivate tumor suppressor genes through suppression
of DNA methyltransferase-1 gene in pancreatic cancer cell lines.
Cancer Biol Ther. 15:419–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Theodore SC, Davis M, Zhao F, Wang H, Chen
D, Rhim J, Dean-Colomb W, Turner T, Ji W, Zeng G, et al: MicroRNA
profiling of novel African American and Caucasian prostate cancer
cell lines reveals a reciprocal regulatory relationship of miR-152
and DNA methyltranferase 1. Oncotarget. 5:3512–3525. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Wu Y, Huang A, Li T, Su X, Ding H, Li H,
Qin X, Hou L, Zhao Q, Ge X, et al: miR-152 reduces human umbilical
vein endothelial cell proliferation and migration by targeting
ADAM17. FEBS Lett. 588:2063–2069. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Su Y, Wang Y, Zhou H, Lei L and Xu L:
MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS
Lett. 588:1983–1988. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Ma J, Yao Y, Wang P, Liu Y, Zhao L, Li Z,
Li Z and Xue Y: miR-152 functions as a tumor suppressor in
glioblastoma stem cells by targeting Krüppel-like factor 4. Cancer
Lett. 355:85–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Cheng Z, Ma R, Tan W and Zhang L: miR-152
suppresses the proliferation and invasion of NSCLC cells by
inhibiting FGF2. Exp Mol Med. 46:e1122014. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Huang S, Xie Y, Yang P, Chen P and Zhang
L: HCV core protein-induced down-regulation of microRNA-152
promoted aberrant proliferation by regulating Wnt1 in HepG2 cells.
PLoS One. 9:e817302014. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Zhai R, Kan X, Wang B, Du H, Long Y, Wu H,
Tao K, Wang G, Bao L, Li F and Zhang W: miR-152 suppresses gastric
cancer cell proliferation and motility by targeting CD151. Tumour
Biol. 35:11367–11373. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Zheng X, Chopp M, Lu Y, Buller B and Jiang
F: miR-15b and miR-152 reduce glioma cell invasion and angiogenesis
via NRP-2 and MMP-3. Cancer Lett. 329:146–154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Zhu C, Li J, Ding Q, Cheng G, Zhou H, Tao
L, Cai H, Li P, Cao Q, Ju X, et al: miR-152 controls migration and
invasive potential by targeting TGFα in prostate cancer cell lines.
Prostate. 73:1082–1089. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Dalmay T and Edwards DR: MicroRNAs and the
hallmarks of cancer. Oncogene. 25:6170–6175. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Lujambio A and Esteller M: CpG island
hypermethylation of tumor suppressor microRNAs in human cancer.
Cell Cycle. 6:1455–1459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Plass C: Cancer epigenomics. Hum Mol
Genet. 11:2479–2488. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Veeck J and Esteller M: Breast cancer
epigenetics: From DNA methylation to microRNAs. J Mammary Gland
Biol Neoplasia. 15:5–17. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Stumpel DJ, Schotte D, Lange-Turenhout EA,
Schneider P, Seslija L, de Menezes RX, Marquez VE, Pieters R, den
Boer ML and Stam RW: Hypermethylation of specific microRNA genes in
MLL-rearranged infant acute lymphoblastic leukemia: Major matters
at a micro scale. Leukemia. 25:429–439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Ragusa M, Majorana A, Banelli B,
Barbagallo D, Statello L, Casciano I, Guglielmino MR, Duro LR,
Scalia M, Magro G, et al: MIR152, MIR200B, and MIR338, human
positional and functional neuroblastoma candidates, are involved in
neuroblast differentiation and apoptosis. J Mol Med (Berl).
88:1041–1053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Chang L and Karin M: Mammalian MAP kinase
signalling cascades. Nature. 410:37–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Rowland BD and Peeper DS: KLF4, p21 and
context-dependent opposing forces in cancer. Nat Rev Cancer.
6:11–23. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Suzuki S, Miyazaki T, Tanaka N, Sakai M,
Sano A, Inose T, Sohda M, Nakajima M, Kato H and Kuwano H:
Prognostic significance of CD151 expression in esophageal squamous
cell carcinoma with aggressive cell proliferation and invasiveness.
Ann Surg Oncol. 18:888–893. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Hoefig KP and Heissmeyer V: MicroRNAs grow
up in the immune system. Curr Opin Immunol. 20:281–287. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Beaulieu AM, Bezman NA, Lee JE, Matloubian
M, Sun JC and Lanier LL: MicroRNA function in NK-cell biology.
Immunol Rev. 253:40–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Manaster I, Goldman-Wohl D, Greenfield C,
Nachmani D, Tsukerman P, Hamani Y, Yagel S and Mandelboim O:
MiRNA-mediated control of HLA-G expression and function. PLoS One.
7:e333952012. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Zhu XM, Han T, Wang XH, Li YH, Yang HG,
Luo YN, Yin GW and Yao YQ: Overexpression of miR-152 leads to
reduced expression of human leukocyte antigen-G and increased
natural killer cell mediated cytolysis in JEG-3 cells. Am J Obstet
Gynecol. 202:592.e1–592.e7. 2010. View Article : Google Scholar
|
|
46.
|
Herrmann TL, Agrawal RS, Connolly SF,
McCaffrey RL, Schlomann J and Kusner DJ: MHC Class II levels and
intracellular localization in human dendritic cells are regulated
by calmodulin kinase II. J Leukoc Biol. 82:686–699. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li
N and Cao X: MicroRNA-148/152 impair innate response and antigen
presentation of TLR-triggered dendritic cells by targeting CaMKIIα.
J Immunol. 185:7244–7251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Nana-Sinkam SP and Croce CM: MicroRNA
dysregulation in cancer: opportunities for the development of
microRNA-based drugs. IDrugs. 13:843–846. 2010.PubMed/NCBI
|
|
49.
|
Mellor HR and Callaghan R: Resistance to
chemotherapy in cancer: a complex and integrated cellular response.
Pharmacology. 81:275–300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Harries M and Gore M: Part I: Chemotherapy
for epithelial ovarian cancer-treatment at first diagnosis. Lancet
Oncol. 3:529–536. 2002. View Article : Google Scholar : PubMed/NCBI
|