Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer worldwide and causes more than half a million
mortalities each year, making it the third most common cause of
cancer-associated mortality (1).
Numerous prognostic factors, including tumor size, tumor quantity,
cell differentiation, venous invasion, degree of inflammation and
deregulated microRNAs (miRNA), have been identified for the
recurrence of HCC, but the molecular mechanism of HCC
carcinogenesis is not fully understood (2–5). In
addition, emerging evidence suggests that components of the RNA
interference pathway and miRNAs may be important in the
carcinogenesis of HCC (6).
miRNAs are small noncoding RNAs, 18–25 nucleotides
in length, which bind to the 3′-untranslated region of target
mRNAs. This sequence-specific binding leads to target mRNA
degradation or repression, resulting in decreased levels of encoded
protein (7,8). miRNAs have already been implicated as
regulators of numerous physiological and pathological processes,
including development, cell proliferation, differentiation and
apoptosis, and tumor progression and metastasis (9). Dicer is a cytoplasmic RNaseIII enzyme
that cleaves the loop of pri-miRNA and long double-stranded RNA
into ~22 bp double-stranded miRNAs and short interfering RNAs. It
is associated with the regulation of cell proliferation and control
of apoptosis (10,11). Increasing evidence has demonstrated
that there is a low expression of Dicer in tumors, including
breast, lung and ovarian cancer, neuroblastoma, human tongue
squamous cell carcinoma, chronic lymphocytic leukemia and
nasopharyngeal carcinoma, and that it is associated with a poor
prognosis as a tumor suppressor (12–23). mRNA
levels of Dicer have been reported to be significantly
downregulated in non-viral HCC patients and this has predicted the
outcome of patients with HCC in previous studies (24,25). The
present study assessed the protein expression levels of Dicer in
HCC patients, the majority of whom had viral HCC, and evaluate its
association with HCC outcome.
Materials and methods
Tissue specimens
HCC tissues were obtained from 119 patients that
underwent HCC resection at The Fourth Hospital of Hebei Medical
University (Shijiazhuang, China) between March 2007 and September
2009. None of the patients had received chemotherapy prior to
surgery. Adjacent non-tumor tissues were used as normal tissues for
comparison to the HCC tissues. Tumor differentiation and
pathological tumor stage were defined by the Edmondson grading
system and the tumor-node-metastasis (TNM) classification of the
International Union Against Cancer (26). The procedures of the present study
were supervised and approved by the Human Tissue Research Committee
of The Fourth Hospital of Hebei Medical University (approval no.,
2007MEC-005), and written informed consent was obtained from all
the patients.
Immunohistochemistry
Serial tissue sections (4 µm thick) were
deparaffinized with xylene, rehydrated, and subjected to microwave
antigen retrieval in citrate buffer (pH, 6.0) for 20 min.
Subsequently, 3% hydrogen peroxide (Invitrogen™; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in methanol was used to quench
endogenous peroxidase activity, followed by incubation with 1%
bovine serum albumin (Pierce™; Thermo Fisher Scientific, Inc.) to
block nonspecific binding. Sections were incubated with rabbit
polyclonal anti-DICER (catalog no., ab93380; dilution, 1:100;
Abcam, Cambridge, UK) overnight at 4°C. Subsequent to rinsing with
phosphate buffer (pH, 7.2) three times, the slides were incubated
with biotinylated secondary anti-rabbit IgG antibody for 20 min at
room temperature and stained with 3,3′-diaminobenzidine (catalog
no., kit-0017; Maixin Biotechnology Development, Co., Ltd., Fuzhou,
China), according to the manufacturer's protocol. All the slides
were counterstained with hematoxylin (Beijing Zhongshan Golden
Bridge Biotechnology Co., Beijing, China), dehydrated and mounted
with a coverslip using a standard medium. The slides were
visualized using the Leica DM IL inverted microscope (Leica
Microsystems GmbH, Wetzlar, Germany). Two independent pathologists,
who were blinded to the study design, were employed to review the
results and score all samples, using the HSCORE as described
previously (17,27). The staining intensity was scored as
follows: 0, no staining; 1, weak staining, light-yellow; 2,
moderate-weak staining, yellow brown; 3, moderate-strong staining,
brown; and 4, strong staining, dark brown). The HSCORE represents
the sum of each of the percentages multiplied by the weighted
intensity of staining as follows: HSCORE = (i + 1) π, where i = 1,
2, 3 and 4, and π varies between 0 and 100%. A score of >100 was
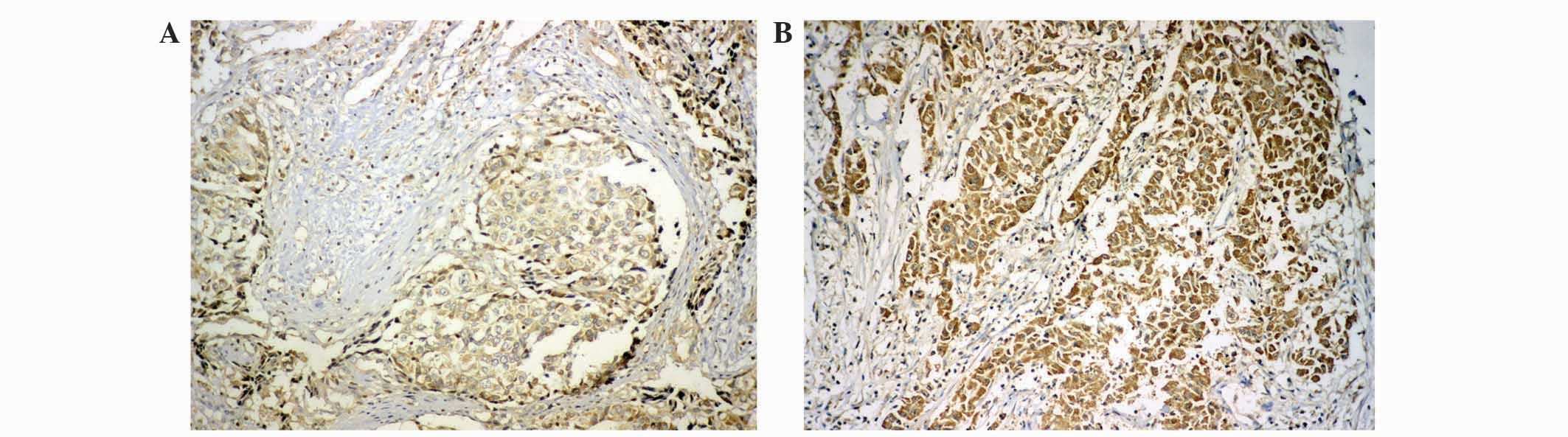
defined as high expression and ≤100 as low expression (Fig. 1).
Cell culture and transfection
Human hepatocellular carcinoma HepG2 cell line was
purchased from the Cell Resource Center, Institute of Basic Medical
Sciences, Chinese Academy of Medical Sciences (Beijing, China). The
cells were maintained as an adherent cell line in Dulbecco's
modified Eagle's medium (DMEM; Gibco®; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum
(Invitrogen™; Thermo Fisher Scientific, Inc.), 1X MEM Non-Essential
Amino Acid, 2 mmol/l L-glutamine, 50 U/ml penicillin and 50 µg/ml
streptomycin (all Gibco®; Thermo Fisher Scientific,
Inc.) in a humidified atmosphere of 5% CO2 at 37°C. The
pcDNA3.1-Dicer vector was a kindly donated by Professor Zhihong
Zheng (China Medical University, Taichung, Taiwan). The HepG2 cells
were transfected with pcDNA3.1 (+) (Invitrogen™; Thermo Fisher
Scientific, Inc.) and pcDNA3.1 (+)-Dicer plasmids using
Lipofectamine® 2000 (Invitrogen™; Thermo Fisher
Scientific, Inc.), which contained geneticin selective markers.
Stable cells were harvested 8 weeks later using geneticin
selection, and the results are represented as PC-control and
PC-Dicer.
Cell proliferation
Cell proliferation capacity was analyzed by Cell
Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) as described previously (28). Briefly, the cells were seeded into
96-well plates (Corning Incorporated, Corning, NY, USA) at a
density of 103 cells/well and incubated for 1–7 days
under the conditions stated previously. In total, 10 µl CCK-8 was
added to each well, according to manufacturer's protocol, followed
by incubation at 37°C with 5% humidified CO2 for 2 h.
The absorbance of the cells was measured at 450 nm with Thermo
Multiskan™ Spectrum microplate reader (Thermo Fisher Scientific,
Inc. CCK-8 allows sensitive colorimetric assays for the
determination of cell proliferation by measuring dehydrogenases in
viable cells. All assays were carried out independently three
times.
Cell migration and invasion assay
Stably transfected cells were harvested and
resuspended in medium without fetal bovine serum and subsequently
placed in the top chamber of a 24-well Transwell plate with 8 µm
pore size (Costar Inc., Milpitas, CA, USA) at a density of
1×105 cells/well. The lower chamber was filled with
media supplemented with 10% fetal bovine serum. After 48 h,
unmigrated cells were removed from the upper surface of the
Transwell membrane using a cotton-tipped swab, and the migrated
cells on the lower membrane surface were fixed with 90% alcohol and
then stained with 0.05% crystal violet (Nanjing Chemical Technology
Co., Ltd., Nanjing, China). The invasion assay was performed in the
same way, with the exception that the membrane insert was coated
with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) diluted in
DMEM (dilution, 1:4). The migrated/invading cells were randomly
counted under a Leica DM IL inverted microscope in 5 representative
areas on each plate (magnification, ×200). The data are presented
as the mean ± standard error of the mean from at least three
independent experiments.
Terminal deoxynucleotidyl transferase
dUTP nick-end labeling (TUNEL) assay
Cell apoptosis was detected using
4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Solarbio
Science and Technology Co., Ltd., Beijing, China) staining and
TUNEL assay. TUNEL assays were performed using the In Situ
Cell Death Detection kit (Roche Diagnostics, Basel, Switzerland),
according to manufacturer's protocol. The cells were fixed in 4%
paraformaldehyde (Nanjing Chemical Technology Co., Ltd.) at room
temperature for 1 h, penetrated with 0.5% Triton X-100 (Amresco
LLC, Solon, OH, USA) for 2 min on ice and then incubated with TUNEL
reaction mixture for 1 h at 37°C. Positive fluorescence was
detected at an excitation wavelength in the range of 570–620 nm
(maximum, 580 nm; red) using a Leica DM IL inverted microscope.
DAPI was excited with ultraviolet light. The experiment was
repeated at least three times.
Western blot analysis
Protein was isolated from cells using RIPA buffer
(Beyotime Institute of Biotechnology, Haimen, China), according to
the manufacturer's instructions. The protein concentration was
measured by performing a BCA assay. Total protein (50 µg) was
denatured by boiling for 5 min and was separated using 7% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis, then
transferred onto a polyvinylidene difluoride membrane
(Sigma-Aldrich, St. Louis, MO, USA). After blocking with 5% skim
milk for 2 h, the membrane was incubated with primary antibody at
4°C overnight. Subsequently, the membrane was incubated with
secondary antibody for 30 min at 37°C and washed by
phosphate-buffered saline with Tween 20. Western blot analysis was
performed with rabbit polyclonal anti-Dicer (dilution, 1:1,000;
catalog no., ab93380; Abcam), rabbit monoclonal anti-B-cell
lymphoma 2 (Bcl-2; dilution, 1:500; catalog no., AB112; Beyotime
Institute of Biotechnology), mouse monoclonal anti-Bcl-2-like
protein 4 (Bax; dilution, 1:500; catalog no., AB026; Beyotime
Institute of Biotechnology) and rabbit polyclonal anti-β-actin
(dilution, 1:500; catalog no., sc-7210; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) antibodies. An HRP-conjugated anti-rabbit
IgG antibody was used as the secondary antibody (dilution, 1:2,000;
catalog no., ZDR-5403; Beijing Zhongshan Golden Bridge
Biotechnology Co.). Signals were detected using
FluorChem® HD2 (Alpha Innotech Corporation, San Leandro,
CA, USA).
Statistical analysis
Survival curves were calculated using the
Kaplan-Meier method, and comparisons between the curves were made
using the log-rank test. Multivariate survival analysis was
performed using a Cox proportional hazards model. Data from the
CCK-8, migration and invasion assay were compared using Student's
t-test. A two-tailed value of P<0.05 was considered to indicate
a statistically significant difference. Statistical analysis was
performed using SPSS version 15.0 software (SPSS, Inc., Chicago,
IL, USA).
Results
Clinical characteristics of patients
with HCC
A total of 119 patients were enrolled in the present
study, which consisted of 32 non-viral HCC patients, 82 hepatitis B
(HBV)-associated HCC patients, 3 hepatitis C (HCV)-associated HCC
patients and 2 HCC patients with HCV and HBV infections. The
patients were followed up every 3 months for 3 years, using routin
radiological and biochemical examination. Survival was measured
between the day of surgery and the date of recurrence, metastasis
and mortality. The association between the data collected during
the 3-year follow-up and the patients' clinical characteristics was
analyzed using the log-rank test with the Kaplan-Meier method.
Gender, age, tumor quantity and child classification (Child-Pugh
grade) (29) were not statistically
significant predictors of postoperative survival time; however,
tumor size, TNM classification and portal vein thrombosis were
associated with survival time in these patients (Table I).
 | Table I.Univariate analysis of clinical
characteristics associated with postoperative survival in patients
with hepatocellular carcinoma. |
Table I.
Univariate analysis of clinical
characteristics associated with postoperative survival in patients
with hepatocellular carcinoma.
| Characteristic | n | 3-year survival rate,
% | P-value |
|---|
| Total | 119 |
|
|
| Gender |
|
|
0.269 |
| Male | 106 | 47.2 |
|
|
Female | 13 | 61.5 |
|
| Age, years |
|
|
0.485 |
| ≤55 | 61 | 45.9 |
|
|
>55 | 58 | 51.7 |
|
| Tumor size, cm |
|
| <0.010 |
| ≤5 | 52 | 69.2 |
|
|
>5 | 67 | 32.8 |
|
| Tumor quantity |
|
|
0.468 |
|
Single | 96 | 50.0 |
|
|
Multiple | 23 | 43.5 |
|
| Child
classification |
|
|
0.258 |
| A | 110 | 50.0 |
|
| B | 9 | 33.3 |
|
| TNM
classification |
|
| <0.010 |
| I | 43 | 74.4 |
|
| II | 70 | 35.7 |
|
|
III | 6 | 16.7 |
|
| Portal vein
thrombosis |
|
| <0.010 |
|
Yes | 16 |
6.3 |
|
| No | 103 | 55.3 |
|
| Dicer
expression |
|
| <0.010 |
|
High | 66 | 63.6 |
|
|
Low | 53 | 30.2 |
|
Dicer expression in HCC tissues, and
its association with HCC survival
Dicer staining was observed in the cytoplasm of
cells in immunohistochemically stained HCC tissues (Fig. 1). A reduced expression of Dicer
(HSCORE 60) was observed in cancerous tissues compared with
adjacent non-cancerous tissues (HSCORE 360). The patients with HCC
were divided into two groups on the basis of Dicer expression
levels: High expression, n=66; low expression, n=53. Postoperative
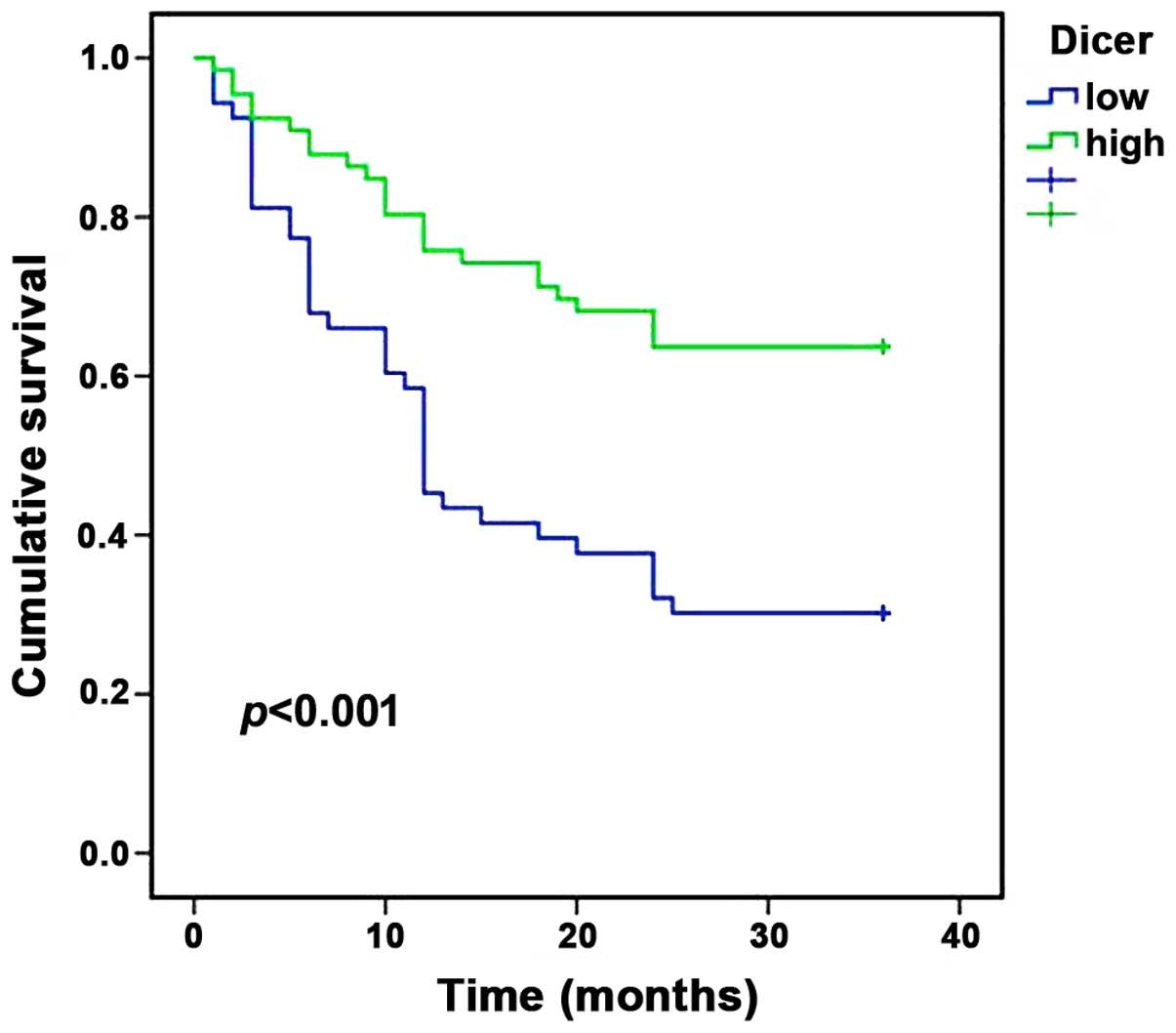
survival curves were plotted using the Kaplan-Meier method for the
high and low Dicer groups, and the results demonstrated that the
3-year survival rates of patients were 63.6% for the high
expression group and 30.2% for the low expression group. Patients
in the low Dicer expression group were associated with a shorter
survival time compared with patients in the high Dicer expression
group (P<0.001; Fig. 2).
Multivariate analysis was performed using Cox
proportional hazards model for various predictive factors. As shown
in Table I, Dicer expression was
identified as an independent predictor of HCC outcome [relative
risk (RR), 0.660; 95% confidence interval, 0.506–0.861; P=0.002].
Size of the tumor (RR, 2.608; 95% CI, 1.429–4.760; P=0.002), portal
vein thrombosis (RR, 2.870; 95% CI, 1.552–5.307; P=0.001), gender
(RR, 2.612; 95% CI, 1.008–6.772; P=0.048) and tumor quantity (RR,
2.870; 95% CI, 1.552–5.307; P=0.001) were also identified as
independent predictive factors for HCC outcome. These data
demonstrate that Dicer expression is a clear predictive factor for
the outcome of HCC patients.
Dicer inhibits proliferation and
induces apoptosis of HCC cells
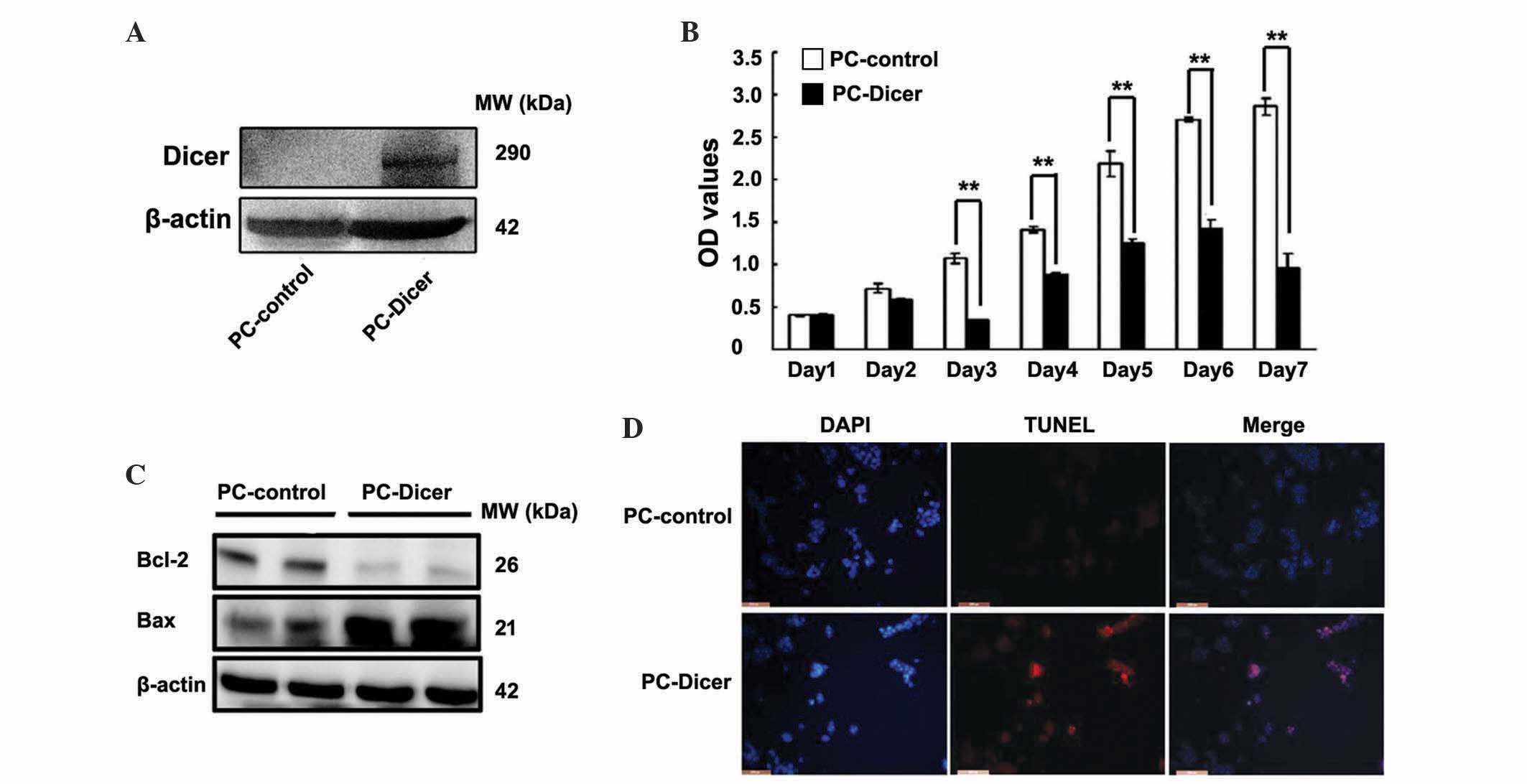
A CCK-8 assay was performed to measure the
proliferation capacity of the HepG2 cells. Cell viability in HepG2
cells overexpressing Dicer was significantly reduced compared with
control cells between day 3 and 7 (Fig.
3A and B; day 1, P=0.077; day 2, P=0.068; day 3, P=0.001; day
4, P=0.004; day 5, P=0.008; day 6, P=0.002; day 7, P=0.001). To
elucidate the potential mechanisms of this growth inhibition by
Dicer, expression of apoptosis-associated factors Bax and Bcl-2
were assessed using western blot analysis, and a TUNEL assay was
performed. Protein levels of Bcl-2 were significantly reduced
whereas Bax protein levels were markedly elevated in the PC-Dicer
HCC cells. The presence of TUNEL-positive cells (stained red) also
revealed that DNA strand breaks occurred in HepG2 cells transfected
with PC-Dicer (Fig. 3C and D). These
data indicate that Dicer inhibits the proliferation and promotes
the apoptosis of HCC cells.
Dicer overexpression suppressed the
migration and invasion of HCC cells
As migration and invasion is an important factor for
tumor metastasis, the effects of Dicer overexpression on cell
migration and invasion was evaluated using Transwell assays. The
results demonstrated that compared with the control cells, PC-Dicer
stably transfected cells had significantly decreased migration
(P<0.01) and invasion (P<0.05) capacities (Fig. 4). Overexpression of Dicer reduced the
migration capacity of cells by ~70% and invasive capacity by ~45%
in Dicer overexpressed HepG2 cells compared with control cells.
Discussion
A reduction of miRNA expression has been identified
in various cancer models using miRNA expression profiling analyses,
which has led to the generation of the hypothesis that miRNA
processing machinery genes, including Dicer, are deregulated in
tumor tissues (30). Low Dicer
expression has been revealed to be associated with advanced tumor
stages and poor clinical outcome in numerous types of cancer
(16–23,31,32).
However, controversial data has been obtained in certain types of
cancer; Dicer has been observed to be upregulated in prostate
adenocarcinoma, ovarian serous carcinoma and colorectal cancer
(33–36).
The present study evaluated the protein level of
Dicer in HCC tissue to investigate its association with HCC
tumorigenesis. Consistent with previous reports that demonstrated
that there is reduced Dicer mRNA expression in nonviral HCC
patients (25), the present study
revealed that there were reduced Dicer protein levels in viral HCC
patients, and that the expression of Dicer was associated with
postoperative HCC survival as a tumor suppressor gene. The
potential mechanism of Dicer in HCC may be explained to a certain
degree by the present functional assay data, which indicated that
Dicer inhibited the proliferation, migration and invasion and
promoted the apoptosis of HCC cells. The present data corroborates
previous observations that Dicer C-terminal fragment possesses
DNase activity that is critical for DNA fragmentation during
apoptosis (37).
A previous study using a miRNA microarray was
performed to identify candidate miRNAs regulated by Dicer using a
small HCC sample size (38). That
study demonstrated that the miRNA expression profiling was
downregulated in tissues that had a low expression of Dicer.
Proliferative and apoptotic (mitogen-activated protein kinases and
Wnt), adhesive (cell adhesion molecules and adherens junction) and
survival (ErbB) signaling pathways appeared to be involved in HCC
tumorigenesis through Dicer mediation (data not shown).
Angiogenesis is a novel target for cancer therapy; however, the
expression of the angiogenesis associated factors vascular
endothelial growth factor and E-cadherin in HepG2 cells were not
altered during Dicer overexpression (39).
In conclusion, the present study demonstrated that
Dicer is important in HCC progression by mediating the
proliferation, apoptosis and metastasis of HCC cells. Therefore,
additional study to understand the mechanism of Dicer in HCC is
crucial in the development of novel methods for the diagnosis and
prognosis of this disease.
Acknowledgements
The present study was supported by Key Basic
Research Program of Hebei (grant nos. 14967713D and 13967607D).
References
|
1
|
Gomaa AI, Khan SA, Toledano MB, Waked I
and Taylor-Robinson SD: Hepatocellular carcinoma: Epidemiology,
risk factors and pathogenesis. World J Gastroenterol. 14:4300–4308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo Z, Wu C, Wang X, Wang C, Zhang R and
Shan B: A polymorphism at the miR-502 binding site in the
3′-untranslated region of the histone methyltransferase SET8 is
associated with hepatocellular carcinoma outcome. Int J Cancer.
131:1318–1322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo Z, Li LQ, Jiang JH, Ou C, Zeng LX and
Xiang BD: Cancer stem cell markers correlate with early recurrence
and survival in hepatocellular carcinoma. World J Gastroenterol.
20:2098–2106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li X, Yang W, Lou L, Chen Y, Wu S and Ding
G: MicroRNA: A promising diagnostic biomarker and therapeutic
target for hepatocellular carcinoma. Dig Dis Sci. 59:1099–1107.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Minagawa M, Makuuchi M, Takayama T and
Kokudo N: Selection criteria for repeat hepatectomy in patients
with recurrent hepatocellular carcinoma. Ann Surg. 238:703–710.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Callegari E, Gramantieri L, Domenicali M,
D'Abundo L, Sabbioni S and Negrini M: MicroRNAs in liver cancer: A
model for investigating pathogenesis and novel therapeutic
approaches. Cell Death Differ. 22:46–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bagga S, Bracht J, Hunter S, Massirer K,
Holtz J, Eachus R and Pasquinelli AE: Regulation by let-7 and lin-4
miRNAs results in target mRNA degradation. Cell. 122:553–563. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuehbacher A, Urbich C, Zeiher AM and
Dimmeler S: Role of Dicer and Drosha for endothelial microRNA
expression and angiogenesis. Circ Res. 101:59–68. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barbato C, Ciotti MT, Serafino A,
Calissano P and Cogoni C: Dicer expression and localization in
post-mitotic neurons. Brain Res. 1175:17–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cochrane DR, Cittelly DM, Howe EN,
Spoelstra NS, McKinsey EL, LaPara K, Elias A, Yee D and Richer JK:
MicroRNAs link estrogen receptor alpha status and Dicer levels in
breast cancer. Horm Cancer. 1:306–319. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dedes KJ, Natrajan R, Lambros MB, Geyer
FC, Lopez-Garcia MA, Savage K, Jones RL and Reis-Filho JS:
Down-regulation of the miRNA master regulators Drosha and Dicer is
associated with specific subgroups of breast cancer. Eur J Cancer.
47:138–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan M, Huang HY, Wang T, Wan Y, Cui SD,
Liu ZZ and Fan QX: Dysregulated expression of Dicer and drosha in
breast cancer. Pathol Oncol Res. 18:343–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Caffrey E, Ingoldsby H, Wall D, Webber M,
Dinneen K, Murillo LS, Inderhaug C, Newell J, Gupta S and Callagy
G: Prognostic significance of deregulated Dicer expression in
breast cancer. PLoS One. 8:e837242013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karube Y, Tanaka H, Osada H, Tomida S,
Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S,
Mitsudomi T and Takahashi T: Reduced expression of Dicer associated
with poor prognosis in lung cancer patients. Cancer Sci.
96:111–115. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Merritt WM, Lin YG, Han LY, Kamat AA,
Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick
AM, et al: Dicer, Drosha, and outcomes in patients with ovarian
cancer. N Engl J Med. 359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Köbel M, Gilks CB and Huntsman DG: Dicer
and Drosha in ovarian cancer. N Engl J Med. 360:1150–1151; author
reply 1151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pampalakis G, Diamandis EP, Katsaros D and
Sotiropoulou G: Down-regulation of Dicer expression in ovarian
cancer tissues. Clin Biochem. 43:324–327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY,
Diccianni MB, London WB, Chang CH and Yu AL: MicroRNA signature and
expression of Dicer and Drosha can predict prognosis and delineate
risk groups in neuroblastoma. Cancer Res. 70:7841–7850. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zeng S, Yang J, Zhao J, Liu Q, Rong M, Guo
Z and Gao W: Silencing Dicer expression enhances cellular
proliferative and invasive capacities in human tongue squamous cell
carcinoma. Oncol Rep. 31:867–873. 2014.PubMed/NCBI
|
|
22
|
Zhu DX, Fan L, Lu RN, Fang C, Shen WY, Zou
ZJ, Wang YH, Zhu HY, Miao KR, Liu P, et al: Downregulated Dicer
expression predicts poor prognosis in chronic lymphocytic leukemia.
Cancer Sci. 103:875–881. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo X, Liao Q, Chen P, Li X, Xiong W, Ma
J, Li X, Luo Z, Tang H, Deng M, et al: The microRNA-processing
enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal
carcinoma. J Cancer Res Clin Oncol. 138:49–56. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu JF, Shen W, Liu NZ, Zeng GL, Yang M,
Zuo GQ, Gan XN, Ren H and Tang KF: Down-regulation of Dicer in
hepatocellular carcinoma. Med Oncol. 28:804–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kitagawa N, Ojima H, Shirakihara T,
Shimizu H, Kokubu A, Urushidate T, Totoki Y, Kosuge T, Miyagawa S
and Shibata T: Downregulation of the microRNA biogenesis components
and its association with poor prognosis in hepatocellular
carcinoma. Cancer Sci. 104:543–551. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Edge SB, Byrd DR and Compton CC: The
American Joint Committee on Cancer: The 7th edition of the AJCC
cancer staging manual and the future of TNM. Ann Surg Oncol.
17:1471–1474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Singh M, Zaino RJ, Filiaci VJ and Leslie
KK: Relationship of estrogen and progesterone receptors to clinical
outcome in metastatic endometrial carcinoma: A Gynecologic oncology
group study. Gynecol Oncol. 106:325–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang M and Zhou H: Grass carp transforming
growth factor-beta 1 (TGF-beta 1): Molecular cloning, tissue
distribution and immunobiological activity in teleost peripheral
blood lymphocytes. Mol Immunol. 45:1792–1798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pugh RN, Murray-Lyon IM, Dawson JL,
Pietroni MC and Williams R: Transection of the oesophagus for
bleeding oesophageal varices. Br J Surg. 60:646–649. 1973.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jafarnejad SM, Ardekani GS, Ghaffari M,
Martinka M and Li G: Sox4-mediated Dicer expression is critical for
suppression of melanoma cell invasion. Oncogene. 32:2131–2139.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuang Y, Cai J, Li D, Han Q, Cao J and
Wang Z: Repression of Dicer is associated with invasive phenotype
and chemoresistance in ovarian cancer. Oncol Lett. 5:1149–1154.
2013.PubMed/NCBI
|
|
33
|
Bian XJ, Zhang GM, Gu CY, Cai Y, Wang CF,
Shen YJ, Zhu Y, Zhang HL, Dai B and Ye DW: Down-regulation of Dicer
and Ago2 is associated with cell proliferation and apoptosis in
prostate cancer. Tumour Biol. 35:11571–11578. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Faber C, Horst D, Hlubek F and Kirchner T:
Overexpression of Dicer predicts poor survival in colorectal
cancer. Eur J Cancer. 47:1414–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Faggad A, Budczies J, Tchernitsa O,
Darb-Esfahani S, Sehouli J, Müller BM, Wirtz R, Chekerov R,
Weichert W, Sinn B, et al: Prognostic significance of Dicer
expression in ovarian cancer-link to global microRNA changes and
oestrogen receptor expression. J Pathol. 220:382–391.
2010.PubMed/NCBI
|
|
36
|
Gao C, Li X, Tong B, Wu K, Liu Y, Anniko M
and Duan M: Up-regulated expression of Dicer reveals poor prognosis
in laryngeal squamous cell carcinoma. Acta Otolaryngol.
134:959–963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakagawa A, Shi Y, Kage-Nakadai E, Mitani
S and Xue D: Caspase-dependent conversion of Dicer ribonuclease
into a death-promoting deoxyribonuclease. Science. 328:327–334.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Krill KT, Gurdziel K, Heaton JH, Simon DP
and Hammer GD: Dicer deficiency reveals microRNAs predicted to
control gene expression in the developing adrenal cortex. Mol
Endocrinol. 27:754–768. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu YY, Chen L, Wang GL, Zhang YX, Zhou JM,
He S, Qin J and Zhu YY: Inhibition of hepatocellular carcinoma
growth and angiogenesis by dual silencing of NET-1 and VEGF. J Mol
Histol. 44:433–445. 2013. View Article : Google Scholar : PubMed/NCBI
|