Introduction
Although the incidence of ovarian cancer ranked
eighth and the mortality ranked seventh in females worldwide in
2008 (1), ovarian cancer remains one
of the most prevalent, deadly and costly gynecological cancers
(2). Epithelial ovarian cancer (EOC)
accounts for almost 90% of all ovarian tumors (3–5) and is
characterized by advanced-stage presentation, multiple organ
metastases, peritoneal dissemination and refractory ascites when
diagnosed (6). Although progress in
early detection technology (transvaginal ultrasound, CA125
examination, computed tomography and BRCA gene mutation detection)
and intervention management (surgical management for borderline
ovarian tumors, ovariectomy for high-risk women) has decreased the
mortality rate in multiple fields, advanced-stage diseases,
particularly those that have metastasized, continue to frequently
progress to recurrent disease and have a poor prognosis, with an
average disease progression-free survival time of 18 months and a
5-year overall survival rate of <30% (7–9).
At present, the diagnosis of metastasis depends on
detection by ultrasound and computed tomography scanning. As a
supplemental diagnostic marker, the level of carbohydrate antigen
125 (CA125) is also considered (10).
However, these measures have limited utility due to their
inadequate sensitivity and specificity (11); the vast majority of metastases
continue to be found during surgery due to false negative tests
(12). Therefore, the development of
novel biomarkers for early detection of EOC with metastasis
(EOC/DM) is imperative and urgent.
There are a large number of non-protein-coding RNAs
in the human genome, which do not encode proteins and were once
considered as ‘noise’ in genome transcription (13,14).
Previous studies have found that these non-protein coding RNAs
regulate cell differentiation, growth and metabolism, in addition
to numerous other biological processes, at the transcriptional,
post-transcriptional or translational levels, and at the DNA, RNA
or protein levels (15–18). Metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) is one of the few well-studied
long non-coding RNAs (lncRNAs), which presents high expression and
enhanced activity in numerous tumor tissues and cells (19–21),
playing a critical role in tumor invasion and metastasis (22). Studies have verified that MALAT1 is
closely associated with metastasis in non-small cell lung cancer
(NSCLC) (23,24) and several other human solid tumors,
including colorectal cancer, osteosarcoma, bladder cancer and
gallbladder cancer (25–28). At present, there is a lack of research
concerning plasma MALAT1 expression in patients with metastatic
EOC.
Materials and methods
Patients and samples
A case-control study was designed to identify
whether MALAT1 could be used as a substitute marker for EOC/DM. All
patients were enrolled from the Xianyang Central Hospital
(Xianyang, Shaanxi, China) between January 2010 and November 2014.
The present study was approved by the Clinical Research Ethics
Committee of Xianyang Central Hospital and all individuals provided
written informed consent. Plasma preparation and RNA isolation were
performed on the clinical samples obtained.
The present cohort totaled 141 female participants,
consisting of 47 patients with EOC/DM, 47 patients with EOC without
metastasis (EOC/NDM) and 47 healthy control (HC) individuals. The
clinical stage of EOCs was conducted according to the
tumor-node-metastasis (TNM) (29) and
Federation of Gynecology and Obstetrics (30) staging systems. The inclusion criteria
for EOC/DM were: Liver, lung or other distant organ metastasis
identified by imaging and confirmed pathologically. The 47 patients
with EOC/NDM had no pathological evidence of distant organ
metastasis; HC individuals were also recruited as patients who
underwent routine medical examination and had no evidence of tumor
disease in the same period. The HC individuals were matched to the
patients based on age and gender. The baseline features of the
patients and HC individuals are summarized in Table I.
 | Table I.Clinicopathological features of EOC
patients and HC individuals. |
Table I.
Clinicopathological features of EOC
patients and HC individuals.
| Variables | EOC/DM, n (%) | EOC/NDM, n (%) | HC, n (%) |
|---|
| Totala | 47
(100.0) | 47
(100.0) | 47 (100.0) |
| Age, years |
|
|
|
|
Median | 58 | 59 | 59 |
|
Range | 49–64 | 48–66 | 48–68 |
| TNM stage |
|
|
|
|
III | 0 (0.0) | 4 (8.5) |
|
| IV | 47
(100.0) | 43 (91.5) |
|
| Pathology
differentiation |
|
|
|
|
Well | 5
(10.6) | 7
(14.9) |
|
|
Moderate | 17 (36.2) | 19 (40.4) |
|
|
Poor | 25 (53.2) | 21 (44.7) |
|
| Serum CA125
level |
|
|
|
| ≤35
U/ml | 2 (4.3) | 3 (6.4) |
|
| >35
U/ml | 45 (95.7) | 44 (93.6) |
|
| Peritoneal
invasion |
|
|
|
|
Absence | 27 (57.4) | 30 (63.8) |
|
|
Presence | 20 (42.6) | 17 (36.2) |
|
| Lymph node
metastasis |
|
|
|
| N1 | 14 (29.8) | 17 (36.2) |
|
| N2 | 20 (42.6) | 17 (36.2) |
|
| N3 | 13 (27.7) | 13 (27.7) |
|
| Metastasis
location |
|
|
|
|
Absence | 0 (0.0) | 4 (8.5) |
|
|
Liver | 29 (61.7) | 28 (59.6) |
|
|
Lung | 11 (23.4) | 10 (21.3) |
|
|
Multiple organ | 7
(14.9) | 5
(10.6) |
|
Plasma preparation
All plasma samples were collected prior to any
therapeutic procedure, such as surgery, chemotherapy or
radiotherapy. Plasma sample preparation and total RNA isolation
were performed as previously described (31). In brief, 5 ml venous blood was
collected in an EDTA gel tube, and the separation procedure was
performed within 2 h of sample collection. Plasma samples were
centrifuged at 1,000 × g for 10 min, followed by 13,000 × g for a
further 10 min at 4°C. The supernatant plasma was then separated,
split into 250 µl aliquots, and frozen at −80°C until use.
RNA preparation, reverse transcription
(RT) and RT-quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from plasma using TRIzol LS
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), following the manufacturer's protocol with minor
modifications (when precipitating RNA, RNase-free glycogen was not
added as a carrier to the aqueous phase). RNA was removed from
genomic DNA and reverse transcribed to cDNA using a
PrimeScript® RT reagent Kit with gDNA Eraser (catalog
no., DRR047; Takara Bio, Inc., Otsu, Shiga, Japan). Subsequently,
RT-qPCR was performed using SYBR® Premix Ex Taq II
(Perfect Real Time; catalog no., DRR081; Takara Bio, Inc.) on
ABI7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.). To
examine the expression of MALAT1 in EOC, RT-qPCR amplification was
performed using the following specific primers: MALAT1 sense,
5′-CTTCCCTAGGGGATTTCAGG-3′ and antisense,
5′-GCCCACAGGAACAAGTCCTA-3′ (32).
β-actin was used as the internal reference, and the primers were as
follows: β-actin sense, 5′-CTTAGTTGCGTTACACCCTTTCTTG-3′ and
antisense, 5′-CTGTCACCTTCACCGTTCCAGTTT-3′. Amplification was
performed under the following conditions: 95°C for 30 sec, followed
by 40 cycles of 95°C for 5 sec, 60°C for 30 sec and 72°C for 30
sec. Melting curve analysis was performed as follows: 95°C for 0
sec (at 20°C/sec from 72–95°C); and 65°C for 15 sec to 95°C for 0
sec (at 0.1°C/sec). Experiments were performed in triplicate in the
same reaction. The results of RT-qPCR experiments were calculated
using the 2−ΔΔCq method (33).
Statistical analysis
Normalized MALAT1 levels were demonstrated as ΔCq,
with ΔCq = Cq (MALAT1) - Cq (β-actin). The difference between
groups was analyzed by the Mann-Whitney or Kruskal-Wallis tests.
Receiver operating characteristic (ROC) curves and the area under
the curve (AUC) were conducted to determine the diagnostic power of
plasma MALAT1 in distant metastasis. COX analysis was used to
assess the clinicopathological factors that were associated with
survival in the EOC patients. The clinicopathological features were
assessed by one-way analysis of variance, Student's t-test or
χ2 test, as appropriate. For the post-hoc test, multiple
comparisons between the groups were performed using the
Student-Newman-Keuls method. P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
conducted using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA).
Graphs and charts were created in GraphPad Prism (version 5.00 for
Windows; GraphPad Software, Inc., La Jolla, CA, USA) and MedCalc
software (version 12.3.0.0; MedCalc Software, Ostend, Belgium).
Results
Evaluation of clinical features in
study groups
The baseline clinicopathological features of the
participants enrolled in the present study are summarized in
Table I. All 94 EOC patients and 47
HC individuals did not differ in age or gender. In addition, there
was no significant difference in the distribution of TNM stage,
pathological differentiation, serum CA125 level, peritoneal
invasion status, lymph node metastasis and metastatic sites between
the EOC/DM and the EOC/NDM groups.
MALAT1 expression was upregulated in
the plasma of metastatic ovarian epithelial carcinoma patients
The expression levels of plasma MALAT1 in 94 EOC
patients and 47 HC individuals were examined by RT-qPCR. The
findings demonstrated that, compared with the EOC/NDM and HC
groups, the expression of plasma MALAT1 was significantly higher in
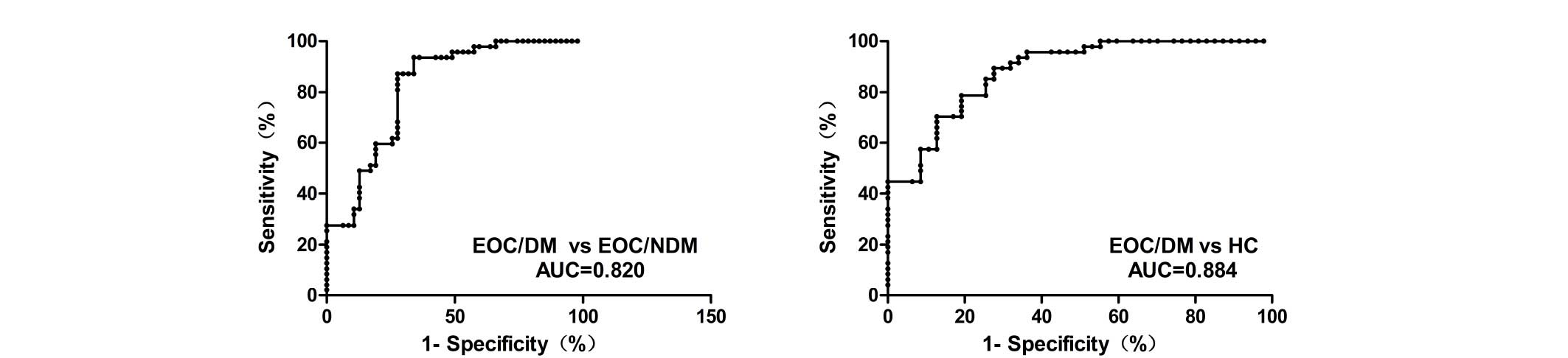
the EOC/DM group (P<0.001 vs. EOC/NDM and HC; Fig. 1). In addition, Youden's index
(sensitivity plus specificity minus 1) was chosen as the optimal
cutoff value and ROC curves were constructed to estimate the
diagnostic value of MALAT1 in discriminating distant metastasis in
EOCs. ROC analyses indicated that plasma MALAT1 yielded an AUC of
0.820 [95% confidence interval (CI), 0.734–0.905; P<0.001], with
87.2% sensitivity and 72.3% specificity for distinguishing EOC/DM
from EOC/NDM. ROC analysis also yielded an AUC of 0.884 (95% CI,
0.820–0.949; P<0.001), with 89.4% sensitivity and 72.3%
specificity for distinguishing EOC/DM from HC. The plasma levels of
MALAT1 were effectively distinguished in patients with EOC/DM from
those with EOC/NDM or HC individuals (Fig. 2).
Cox analysis and Kaplan-Meier survival
analysis of prognostic factors in EOC
Effect on the prognosis of plasma MALAT1 expression
and other clinicopathological factors was also assessed. Low levels
of CA125 were considered as ≤35 U/ml, while high levels of CA125
were >35 U/ml. Low levels of plasma MALAT1 were considered as a
fold-change of <2, while high levels of plasma MALAT1 were a
fold-change of >2. Univariate and multivariate Cox analysis
revealed that poor differentiation, TNM stage IV, N3 lymph node
metastasis, presence of peritoneal invasion, high serum CA125
levels and high plasma MALAT1 levels were significantly associated
with short life expectancy (Table
II). Using the Kaplan-Meier and log-rank methods, it was found
that high expression of the plasma MALAT1 was associated with a
shorter overall survival (OS) of patients with EOC (hazard ratio,
3.322; 95% CI, 1.140–5.680; P=0.028; Fig.
3). Upregulation of plasma MALAT1 may independently contribute
to a poor prognosis in patients with EOC.
 | Table II.Univariate and multivariate analysis
of plasma MALAT1 and clinicopathological factors associated with
survival in epithelial ovarian cancer patients. |
Table II.
Univariate and multivariate analysis
of plasma MALAT1 and clinicopathological factors associated with
survival in epithelial ovarian cancer patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Differentiation
(poor) | 3.576 | 1.356–6.754 | 0.015 | 2.357 | 1.085–5.867 | 0.023 |
| TNM stage (IV) | 2.376 | 0.989–5.374 | 0.005 | 1.864 | 0.956–4.356 | 0.043 |
| Metastasis location
(lung) | 1.137 | 0.376–3.202 | 0.082 | – | – | – |
| Lymph node
metastasis (N3) | 1.271 | 0.352–2.328 | 0.043 | 1.479 | 0.561–4.546 | 0.037 |
| Peritoneal invasion
(presence) | 2.725 | 0.934–6.347 | 0.025 | 2.435 | 0.834–4.563 | 0.039 |
| Serum CA125 (>35
U/ml) | 2.259 | 0.563–3.194 | 0.018 | 3.189 | 1.043–6.487 | 0.017 |
| Plasma MALAT1
(high) | 2.391 | 1.057–4.672 | 0.031 | 3.322 | 1.140–5.680 | 0.028 |
Discussion
EOC ranks among the most common and lethal malignant
diseases in gynecological cancers. Due to the location of the
ovaries and the biological characteristics of insidious onset of
EOC, it becomes extremely challenging to diagnose ovarian cancer at
an early curable stage. In total, >70% of patients present with
advanced disease at diagnosis (34).
The cure rate of women with advanced ovarian cancer is ≤40% as a
result of challenging and inefficient surgery on metastatic disease
(35). Poor prognosis of EOC is
associated with the early onset of tumor metastasis. The mechanisms
of EOC metastasis development are poorly understood and require
additional exploration. Several studies have found that abnormal
expression of lncRNAs are involved in tumor metastasis through
regulating the expression of other molecules (36–39). Ji
et al (22) identified the
MALAT1 lncRNA by subtractive hybridization, establishing it as a
prognostic marker for metastasis and survival in NSCLC. However,
the roles of MALAT1 in the metastasis of EOC have yet to be fully
elucidated.
Due to the features of lncRNAs, such as long
fragments (>200 nt), easy degradation in plasma, and extremely
low concentration of total RNA in plasma, the current detection of
plasma lncRNA as tumor markers becomes extremely challenging. Arita
et al found that the lncRNA full-length form is not stable
in plasma (40), but certain
fragments in the plasma are highly stable and abundant (32,41,42). The
present study assessed the relevance of plasma MALAT1 to metastasis
of EOC. The results demonstrated that a specific stable MALAT1
fragment existed in plasma, which was identified by a previous
study (43), thus making circulating
lncRNA expression detection available.
The present results showed that the level of plasma
MALAT1 was markedly increased in patients with EOC/DM compared with
patients with EOC/NDM and the HC group, and could effectively
identify patients in the EOC/DM group from those in the EOC/NDM and
HC group, thus indicating a suitable plasma biomarker for MALAT1 in
EOC metastasis development. Furthermore, through Kaplan-Meier
survival analysis and Cox analysis, it was found that elevated
MALAT1 expression was associated with a shorter OS time in patients
with EOC, as well as poor differentiation, advanced-stage disease,
multiple lymph node metastases, presence of peritoneal invasion and
increased serum CA125 level. Although the mechanism remains to be
elucidated, these findings indicated that MALAT1 may play an
important role in the cancer metastatic process, and may be a
useful novel marker for metastatic EOC. Similar to the present
findings, Shen et al (44)
found that the level of MALAT1 was significantly increased in brain
metastasis compared with non-brain metastasis samples, and
subsequent functional studies indicated that MALAT1 promoted lung
cancer cell brain metastasis by inducing epithelial-mesenchymal
transition. Several studies have revealed that there are other
MALAT1-mediated molecular pathways involved in tumor metastasis,
such as through binding to splicing factor, proline- and
glutamine-rich (SFPQ) and releasing the oncogene polypyrimidine
tract binding protein 2 (PTBP2) from the SFPQ/PTBP2 complex
(25), activating the extracellular
signal-regulated kinase/Mitogen-activated protein kinase or
phosphoinositide 3-kinase/AKT pathway to result in distant
metastasis in numerous cancer types (26,28).
Overall, these mechanisms may explain why the present patients with
high expression of MALAT1 expression demonstrated a strong
metastasis tendency.
In summary, the high heterogeneity of metastatic EOC
leads to resistance to chemotherapy drugs, poor prognosis and lack
of effective targeting therapy. Therefore, identifying specific EOC
metastasis-associated markers, and subsequently executing an early
intervention have become the ‘bottleneck’ problem of EOC treatment.
In the present study, it was found that plasma MALAT1 upregulation
was closely associated with EOC distant metastasis and may be an
independent risk factor for poor prognosis. The results of the
present study may aid in understanding the molecular mechanisms of
EOC with metastasis, which possess potential clinical value for
metastatic EOC screening and early diagnosis, and may be a
potential target for the treatment of EOC with metastasis.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
MALAT1
|
metastasis-associated lung
adenocarcinoma transcript 1
|
|
EOC
|
epithelial ovarian cancer
|
|
EOC/DM
|
epithelial ovarian cancer with
metastasis
|
|
EOC/NDM
|
epithelial ovarian cancer without
metastasis
|
|
HC
|
healthy controls
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
CA125
|
carbohydrate antigen 125
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan JK, Cheung MK, Husain A, Teng NN,
West D, Whittemore AS, Berek JS and Osann K: Patterns and progress
in ovarian cancer over 14 years. Obstet Gynecol. 108:521–528. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prat J: New insights into ovarian cancer
pathology. Ann Oncol. 23(Suppl 10): x111–x117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jelovac D and Armstrong DK: Recent
progress in the diagnosis and treatment of ovarian cancer. CA
Cancer J Clin. 61:183–203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herzog TJ and Pothuri B: Ovarian cancer: A
focus on management of recurrent disease. Nat Clin Pract Oncol.
3:604–611. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Greenlee RT, Hill-Harmon MB, Murray T and
Thun M: Cancer statistics, 2001. CA Cancer J Clin. 51:15–36. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoskins P, Eisenhauer E, Vergote I,
Dubuc-Lissoir J, Fisher B, Grimshaw R, Oza A, Plante M, Stuart G
and Vermorken J: Phase II feasibility study of sequential couplets
of Cisplatin/Topotecan followed by paclitaxel/cisplatin as primary
treatment for advanced epithelial ovarian cancer: A national cancer
institute of canada clinical trials group study. J Clin Oncol.
18:4038–4044. 2000.PubMed/NCBI
|
|
10
|
Theriault C, Pinard M, Comamala M,
Migneault M, Beaudin J, Matte I, Boivin M, Piché A and Rancourt C:
MUC16 (CA125) regulates epithelial ovarian cancer cell growth,
tumorigenesis and metastasis. Gynecol Oncol. 121:434–443. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dorigo O and Berek JS: Personalizing CA125
levels for ovarian cancer screening. Cancer Prev Res (Phila).
4:1356–1359. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakajo M, Nakajo M, Nakayama H, Jinguji M,
Nakabeppu Y, Higashi M, Nakamura Y, Sato M and Yoshiura T:
Dexamethasone suppression FDG PET/CT for differentiating between
true- and false-positive pulmonary and mediastinal lymph node
metastases in non-small cell lung cancer: A pilot study of FDG
PET/CT after oral administration of dexamethasone. Radiology.
279:246–253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fujimoto A, Furuta M, Totoki Y, Tsunoda T,
Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, et
al: Whole-genome mutational landscape and characterization of
noncoding and structural mutations in liver cancer. Nat Genet.
48:500–509. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen D, Sun Q, Cheng X, Zhang L, Song W,
Zhou D, Lin J and Wang W: Genome-wide analysis of long noncoding
RNA (lncRNA) expression in colorectal cancer tissues from patients
with liver metastasis. Cancer Med. May 11–2016.(Epub ahead of
print). View
Article : Google Scholar
|
|
15
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao B, Song N, Zhang M, Di C, Yang Y, Lu
Y, Chen R, Lu ZJ and Guo M: Systematic study of novel lncRNAs in
different gastrointestinal cancer cells. Discov Med. 21:159–171.
2016.PubMed/NCBI
|
|
18
|
Wang L, Li J, Zhao H, Hu J, Ping Y, Li F,
Lan Y, Xu C, Xiao Y and Li X: Identifying the crosstalk of
dysfunctional pathways mediated by lncRNAs in breast cancer
subtypes. Mol Biosyst. 12:711–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
20
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen L, Chen L, Wang Y, Jiang X, Xia H and
Zhuang Z: Long noncoding RNA MALAT1 promotes brain metastasis by
inducing epithelial-mesenchymal transition in lung cancer. J
Neurooncol. 121:101–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding
Q, Weng H, Shu YJ, Liu TY, Jiang L, et al: MALAT1 promotes the
proliferation and metastasis of gallbladder cancer cells by
activating the ERK/MAPK pathway. Cancer Biol Ther. 15:806–814.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
National Comprehensive Cancer Network.
NCCN Clinical Practice Guidelines in Oncology: (NCCN
Guidelines®) Ovarian Cancer Including Fallopian Tube
Cancer and Primary Peritoneal Cancer. Version 1. 2015.http://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdfAccessed.
May 5–2016
|
|
30
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: Ovary and primary peritoneal carcinoma.
AJCC Cancer Staging Manual (7th). Springer-Verlag. (NY). 419–428.
2010.
|
|
31
|
Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang
Q, Chen L, Pang X, Leng W and Bi F: Plasma miR-122 and miR-192 as
potential novel biomarkers for the early detection of distant
metastasis of gastric cancer. Oncol Rep. 31:1863–1870.
2014.PubMed/NCBI
|
|
32
|
Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J,
Wei M, Xu C, Wu C, Zhang Z, et al: Long non-coding RNA metastasis
associated in lung adenocarcinoma transcript 1 derived miniRNA as a
novel plasma-based biomarker for diagnosing prostate cancer. Eur J
Cancer. 49:2949–2959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Barakat RR, Markman M and Randall M:
Principles and Practice of Gynecologic Oncology (5th). Lippincott
Williams & Wilkins. 2009.
|
|
35
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin
H, Zhang H, Zhang H, Liu J, Guo H, et al: Hypoxia-inducible
lncRNA-AK058003 promotes gastric cancer metastasis by targeting
γ-synuclein. Neoplasia. 16:1094–1106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng
R, Wang Y, Huang J, Xu M, Yan J and Yu J: lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting TGFBI. FEBS J.
281:3766–3775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Arita T, Ichikawa D, Konishi H, Komatsu S,
Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T,
et al: Circulating long non-coding RNAs in plasma of patients with
gastric cancer. Anticancer Res. 33:3185–3193. 2013.PubMed/NCBI
|
|
41
|
Tsui NB, Ng EK and Lo YM: Stability of
endogenous and added RNA in blood specimens, serum and plasma. Clin
Chem. 48:1647–1653. 2002.PubMed/NCBI
|
|
42
|
Ng EK, Tsui NB, Lau TK, Leung TN, Chiu RW,
Panesar NS, Lit LC, Chan KW and Lo YM: mRNA of placental origin is
readily detectable in maternal plasma. Proc Natl Acad Sci USA.
100:4748–4753. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Weber DG, Johnen G, Casjens S, Bryk O,
Pesch B, Jöckel KH, Kollmeier J and Brüning T: Evaluation of long
noncoding RNA MALAT1 as a candidate blood-based biomarker for the
diagnosis of non-small cell lung cancer. BMC Res Notes. 6:5182013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shen L, Chen L, Wang Y, Jiang X, Xia H and
Zhuang Z: Long noncoding RNA MALAT1 promotes brain metastasis by
inducing epithelial-mesenchymal transition in lung cancer. J
Neurooncol. 121:101–108. 2015. View Article : Google Scholar : PubMed/NCBI
|