Introduction
Activating transcription factor 3 (ATF3) is a member
of the ATF/cyclic AMP-responsive element binding protein
(CREB)family of transcription factors and has been demonstrated to
form dimers with other ATF/CREB proteins, including c-Jun, ATF2,
Jun D and Jun B (1). As a
transcription factor, ATF3 is critical in cell growth, apoptosis
and carcinogenesis (1). Notably, ATF3
has been reported to serve controversial roles in oncogenesis and
tumor suppression depending on the context and cell type. Several
studies support the oncogenic role of ATF3. For example, ATF3 has
been reported to be overexpressed in breast cancer (2), prostate cancer (3) and Hodgkin lymphomas (4), and high expression is an indicator of
poor prognosis of patients with prostate cancer (3). In vivo and in vitro
studies have also demonstrated that overexpression of ATF3 promotes
cancer cell proliferation and metastasis in prostate cancer
(5,6),
and is associated with upregulation of Slug, fibronectin-1 and
TWIST1 transcripts, which are important regulators of
cell-extracellular matrix or cell-cell interactions (5,6). In
addition, ATF3 overexpression results in the binding of ATF3 to the
GADD153 promoter, which subsequently represses its transcription in
cervical cancer HeLa cells, providing a potential pathway through
which ATF3 is able to promote cancer cell survival (7). In contrast to the aforementioned
studies, growing evidence suggests that ATF3 is able to suppress
tumorigenesis. For example, it has been demonstrated that ATF3
expression levels are reduced in human colorectal cancer, and
overexpression of ATF3 exhibits tumor suppressive roles, such that
the protein reduces metastatic potential and promotes apoptosis in
various cell lines to inhibit carcinogenesis (8,9). In
addition, ATF3 is able to suppress the oncogenic function of mutant
p53 in lung cancer (10). The
possible role of ATF3 as a tumor suppressor is supported by its
established role in transforming growth factor-β (TGF-β) signaling
(11). TGF-β is a potent tumor
suppressor in epithelial cells that signals via Smad3 activation to
directly induce ATF3 (11). Smad3 and
ATF3 subsequently form a complex, which binds to the promoter of
inhibitor of DNA binding 1 (ID1) and directly mediates its
repression (11). Furthermore, ATF3
may be activated by a range of anticancer compounds, including
non-steroidal anti-inflammatory drugs, curcumin, dietary compounds
resveratrol and genistein, progesterone and the
phosphatidylinositol inhibitor LY294002 (1,12).
Inversely, resveratrol and genistein also suppress ID1 expression
(12).
ID1 is a member of the ID protein superfamily, which
belongs to the helix-loop-helix transcription factor family
(13,14). ID1 is ubiquitously expressed in a
number of tissues and functions in a wide range of cellular
processes, including proliferation, cell differentiation,
senescence and apoptosis (15).
Growing evidence suggests that ID1 is an oncogene and is critical
in promoting tumor invasion and development, as it is overexpressed
in human cancer of the pancreas, thyroid, breast, cervix, ovary,
prostate, esophagus and lung, and high expression of ID1 is
associated with a poor prognosis (16–18).
Furthermore, ID1 is able to promote cell survival and induce cancer
cell growth, which may be associated with ATF3 (19).
Recent studies have reported that ATF3 was
downregulated in esophageal squamous cell carcinoma (ESCC) compared
with paired non-cancerous tissues, and that lower ATF3 expression
in tumors was significantly correlated with shorter survival time
(20,21). Furthermore, increased expression of
ATF3 inhibited ESCC cell growth and invasion in vitro and in
nude mice via p53 signaling (21).
However, it is unclear whether ESCC tumor inhibition by ATF3 occurs
through ID1 repression. ESCC is one of the most common malignancies
in worldwide. There were about 477,900 new cases and 375,000 death
of ESCC in China (22). The present
study aimed to determine the association between ATF3 and ID1 in
ESCC tissues and in vitro by manipulating ATF3
expression.
Materials and methods
Human samples and immunohistochemical
staining
A total of 36 pairs of ESCC tissues and their
adjacent non-cancer tissues were obtained from the Tissue Bank of
the Laboratory for Cancer Signal Transduction, Xinxiang Medical
University (Xinxiang, China). All procedures were approved by the
Institutional Review Board of Xinxiang Medical University.
Immunohistochemical staining was conducted as
previously described (23). In brief,
formalin-fixed, paraffin-embedded tissues were sectioned and
deparaffinized with xylene, rehydrated with gradient ethanol and
distilled water, and were subsequently blocked with serum and
incubated with anti-AFT3 (1:100, ca no. ab191513) and anti-ID1
(1:100, cat no. ab134163) primary antibodies (Abcam, Cambridge, MA,
USA) at 4°C overnight. The sections were then incubated with a
biotinylated goat anti-rabbit IgG antibody, dilution (1:200; cat
no. PI-1000; Vector Laboratories, Inc., Burlingame, CA, USA), and a
VECTASTAIN Elite ABC kit (Vector Laboratories, Inc., Burlingame,
CA, USA) was used according to the manufacturer's protocol. The
sections were finally stained with 3,3′-diaminobenzidine.
Immunohistochemical staining was evaluated based on immunostaining
intensities (absent or weak, moderate and strong) as previously
described (23).
Cell culture and transfection
Human ESCC EC109 and KYSE450 cell lines obtained
from American Type Culture Collection (Manassas, VA, USA) were
cultured in Dulbecco's modified Eagle's medium containing 10% fetal
calf serum (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). All cells were incubated at 37°C in a humidified
atmosphere containing 5% CO2. An overexpression plasmid
for ATF3 was constructed as follows: Human ATF3 cDNA was amplified
and inserted into a pcDNA3-Flag vector. The expression plasmid
(pFlag-ATF3) was transfected into the ESCC cell lines using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. An empty
vector (pFlag-cDNA3) was also transfected into the ESCC cells,
which served as a negative control.
Cell proliferation assay
3-(4,5-dimethylthiazol-2-yl)
5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)
assay was performed to determine cytotoxicity of the EC109 and
KYSE450 cells following transfection with pFlag-ATF3 using
Lipofectamine 2000. pFlag-cDNA3 was used as a control for
comparison. Following 24, 36 and 48 h, cell proliferation was
determined by MTS assay using the CellTiter 96®
Non-Radioactive Cell Proliferation Assay kit (Promega Corporation,
Madison, WI, USA) according to the manufacturer's protocol. The
remaining viable cells with MTS uptake were determined by measuring
the optical density at 570 nm using an enzyme-linked immunosorbent
assay reader (Molecular Devices, LLC, Sunnyvale, CA, USA). Values
are presented as the mean ± standard deviation. At least three
measurements were read, and the experiments were conducted three
times independently.
Wound healing and transwell
assays
As previously described (24), EC109 and KYSE450 cells transiently
transfected with pFlag-ATF3 plasmid or the pFlag-cDNA3 empty vector
were seeded in a 100-mm Petri dish. A wound was made by scratching
on the bottom of the dish, followed by a 36 h incubation at 37°C.
The wound healing status was checked under an inverse microscope.
Cell migration was detected using a Transwell plate (Corning Life
Sciences, Lowell, MA, USA). Approximately 1×103 EC109
cells were transiently transfected with the pFlag-ATF3 plasmid or
the pFlag-cDNA3 empty vector, and were subsequently seeded into the
Transwell plate. The cells were cultured in Dulbecco's modified
Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal calf serum (Invitrogen,
Carlsbad, CA). The migrated cells were stained with
4′,6-diamidino-2-phenylindole and counted under an inverse
fluorescence microscope at 48 h post-seeding.
Immunoblotting
For immunoblotting, human ESCC EC109 and KYSE459
cells were collected 48 h post-transfection with pFlag-ATF3 or
pFlag-cDNA3 empty vector. Cells were lysed using 1X
radioimmunoprecipitation assay (RIPA) buffer (Upstate
Biotechnology, Inc., Lake Placid, NY, USA) containing a protease
inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). Following
cell lysis, 45 µg protein was loaded on a 10% sodium dodecyl
sulfate (SDS) gel followed by transfer to a polyvinylidene fluoride
membrane. The membrane was probed with antibodies against Flag (cat
no. ab49763, dilution 1:1000), ATF3 (cat no. ab191513, dilution
1:250), ID1 (cat no. ab134163, 1:250), cyclin D1 (cat no. ab134175,
dilution 1:250), E-cadherin (cat no. ab40772, dilution 1:300;
Abcam), phosphorylated (p)-STAT3 (cat no. 9131s, dilution 1:300),
Twist (cat no. 4119s, 1:300; Cell Signaling Technology, Inc.,
Danvers, MA, USA), glyceraldehyde 3-phosphate dehydrogenase (cat
no. G8795, dilution 1:500) and β-actin (cat no. A8481, dilution
1:1000; Sigma-Aldrich). Secondary antibodies (cat no. SC-2004, goat
anti-rabbit IgG-HRP, dilution 1:1000; cat no. SC-2005, goat
anti-mouse IgG-HRP, dilution 1:1000) were purchased from Santa Cruz
Biotechnolog, Dallas, TX, USA. The signals were visualized using an
enhanced chemiluminescence kit (Beyotime Institute of
Biotechnology, Haimen, China) according to the manufacturer's
protocol.
Quantitative reverse transcriptional
PCR (RT-qPCR) assay
To determine the alterations of ID1 at mRNA levels,
the EC109 and KYSE450 cells transfected with pFlag-ATF3 or
pFlag-cDNA3 were collected after 48 h post transfection. Total RNA
were extracted and RT-qPCR were conducted to determine the changes
of ID1 mRNA using the following primers: ID1-forward:
TGGATGGCGGGTTTCAGATG, ID1-reverse: TCTTCGGTCAGACGATTGACA. The
detailed procedure was reported previously (23).
Co-immunoprecipitation assay
EC109 cells were transfected with pFlag-ATF3 or
pFlag-cDNA3 for 48 h. The cells were collected and incubated on ice
for 15 min in RIPA lysis buffer supplemented with protease
inhibitor cocktail. Total cell lysate was centrifuged at 8,000 × g
for 15 min at 4°C. A total of 300 mg supernatant was incubated with
anti-Flag antibody (GenScript Corporation, Scotch Plains, NJ, USA)
overnight at 4°C on a rotator, followed by the addition of Protein
A/G PLUS-Agarose (Santa Cruz Biotechnology, Inc.) for 2 h at 4°C.
The immunocomplex was separated by 12% SDS-polyacrylamide gel
electrophoresis. The PVDF membrane was probed with anti-ID1
antibody to detect the ATF3/ID1 complex.
Statistical analysis
Data were presented as mean ± standard deviation
(SD), the Student-t test was used for groups' quantity comparison,
and Chi-square test was used for correlation analysis. P<0.05
was considered to indicate a statistically significant
difference.
Results
Inverse correlation between ATF3 and
ID1 expression in the ESCC tissues
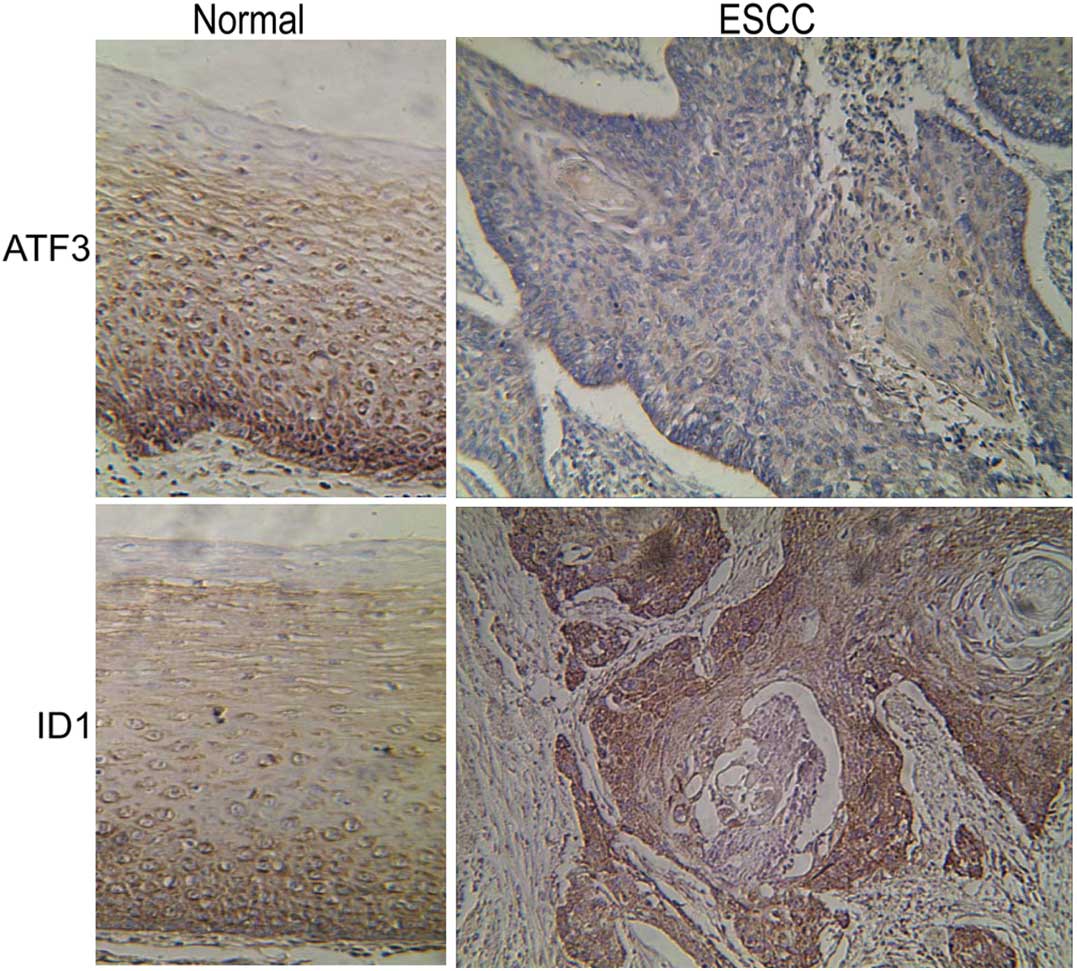
Using immunohistochemical staining, it was observed
that ATF3 was expressed in the ESCC and adjacent non-tumor tissues;
however, ATF3 expression was reduced in the ESCC tissues compared
with the adjacent non-tumor tissues (Fig.
1). By contrast, ID1 was overexpressed in ESCC tissues compared
with the adjacent non-tumor tissues (Fig.
1). These results therefore demonstrate a significant inverse
correlation between ATF3 and ID1 expression in the ESCC tissues
(Table I; X2=9.84;
P<0.01).
 | Table I.Expression status of ATF3 and ID1 and
their correlation in esophageal squamous cell carcinoma
tissues. |
Table I.
Expression status of ATF3 and ID1 and
their correlation in esophageal squamous cell carcinoma
tissues.
| Protein | Absent/weak, n
(%) | Moderate, n
(%) | Strong, n (%) | Total, n (%) |
|---|
| ATF3 | 10 (27.7) | 17 (47.2) | 9
(25.0) | 36 (100.0) |
| ID1 | 4
(11.1) | 10 (27.8) | 22 (61.1) | 36 (100.0) |
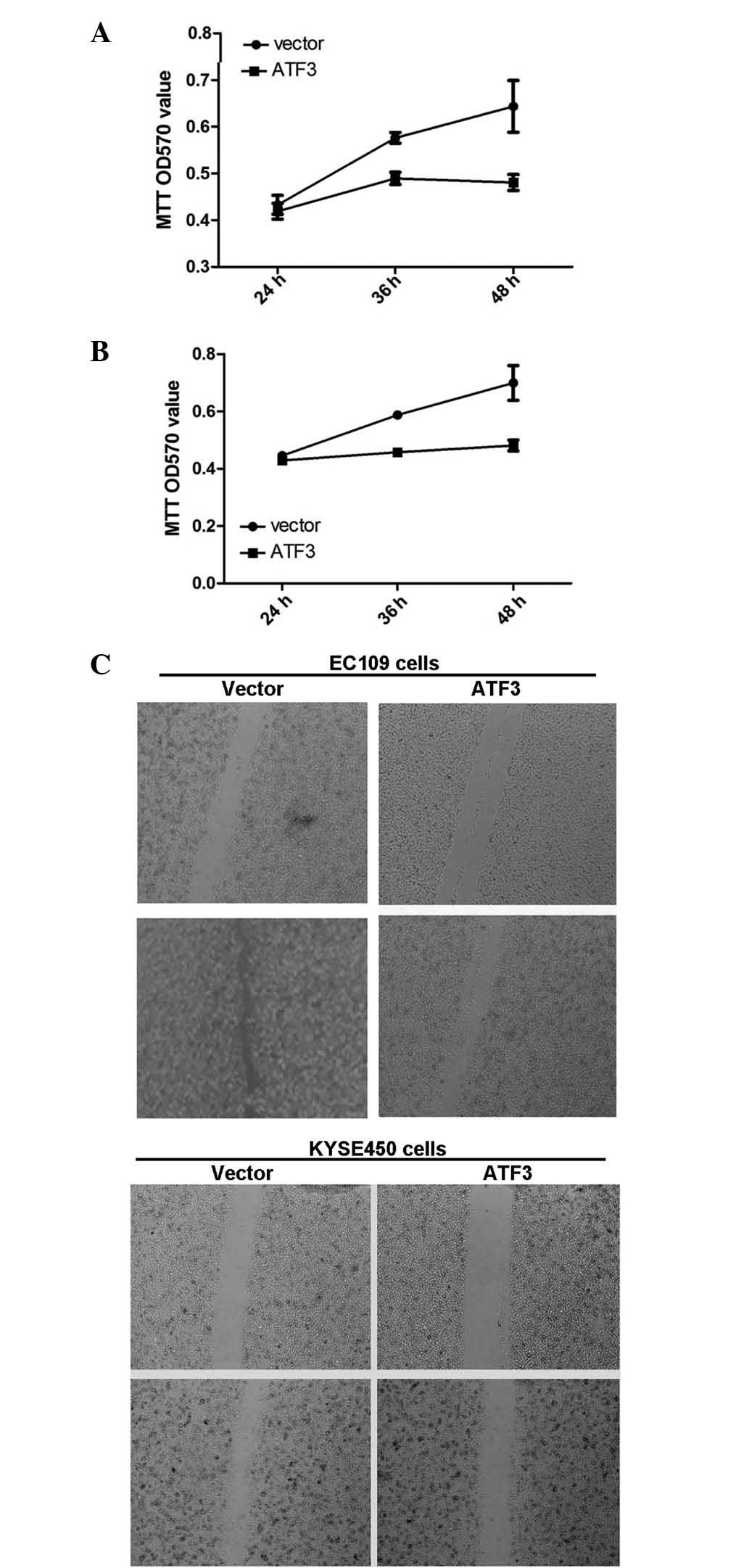
ATF3 inhibits cell proliferation,
motility and migration in ESCC cells
To determine the roles of ATF3 in ESCC cells, the
ATF3 overexpression plasmid was transfected in to the EC109 and
KYSE450 cells. It was observed that increased ATF3 expression
significantly inhibited cell proliferation in each ESCC cell line
(Fig. 2A and B). Wound healing assay
demonstrated that ATF3 overexpression inhibited ESCC cell motility
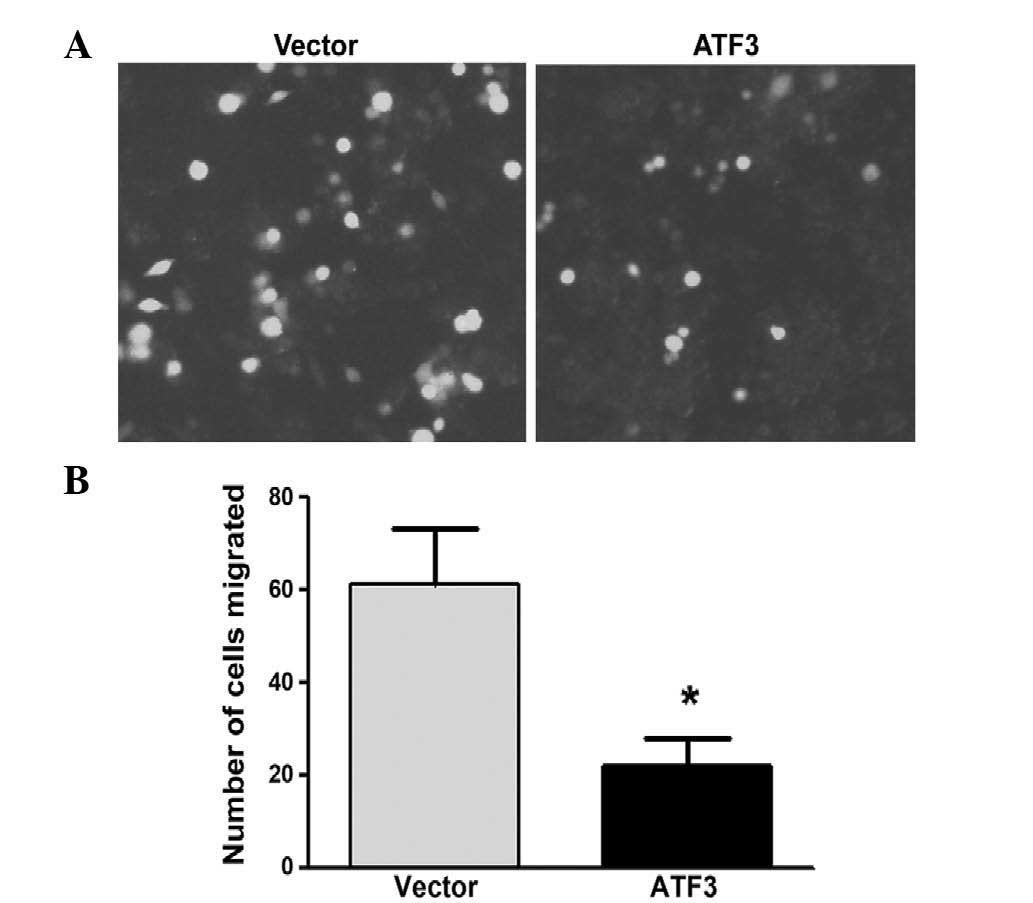
(Fig. 2C), and Transwell assay
determined that ATF3 significantly inhibited ESCC cell migration
(P<0.05) (Fig. 3).
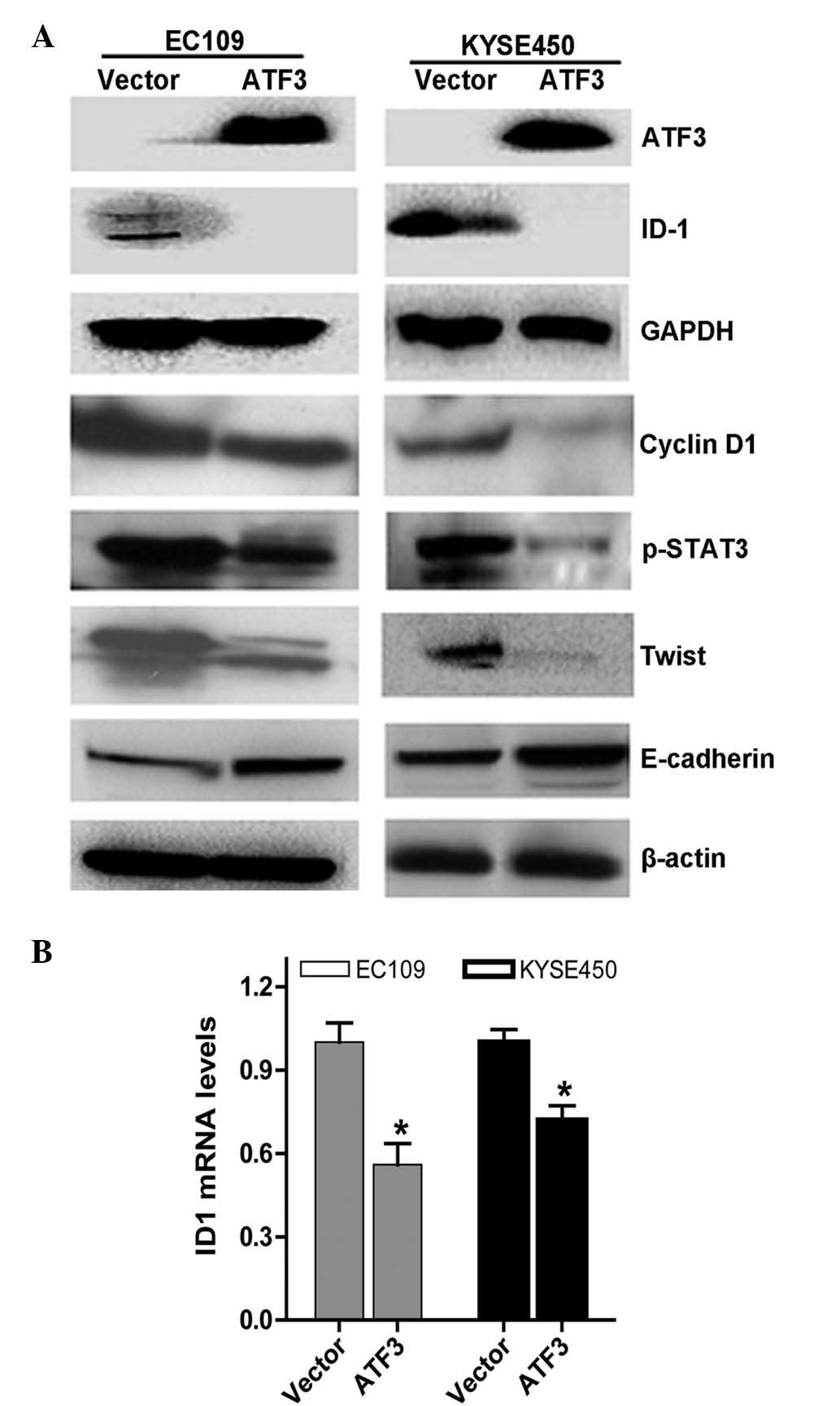
To determine the mechanisms underlying these
results, immunoblotting was performed. The results demonstrated
that ATF3 repressed ID1 expression. In addition, ATF3 downregulated
the expression of cyclin D1, p-STAT3 and TWIST, and upregulated the
expression of E-cadherin (Fig.
4A).
ATF3 represses ID1 expression in ESCC
cells
The present study subsequently investigated the
effects of ATF3 on ID1 expression by transfecting ATF3
overexpression plasmids into ESCC EC109 and KYSE450 cells. The ID1
mRNA levels and their interactions were subsequently determined,
and the results demonstrated that increased expression of ATF3
significantly suppressed ID1 expression at the mRNA level (Fig. 4B) as determined by reverse
transcription-quantitative polymerase chain reaction, which was
consistent with the inhibition of ID1 at the protein level
(Fig. 4A).
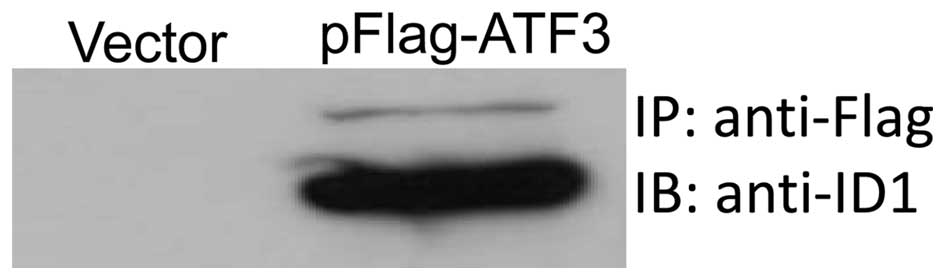
To further investigate the interaction between ATF3
and ID1, co-immunoprecipitation assay was performed. Anti-Flag
antibodies were used to pull down the immune-complex and anti-ID1
antibodies were used to probe the complex. As presented in Fig. 5, there was a strong band in the
precipitated immune-complex, therefore suggesting that the ATF3 and
ID1 proteins had bound together.
Discussion
Esophageal cancer is one of the most prevalent forms
of cancer worldwide and is frequently fatal, with a 5-year survival
rate of <20% (25). The underlying
mechanisms of carcinogenesis and progression remain largely
unknown. Recent studies using genome-wide analysis and whole genome
sequencing have identified several signal nucleotide polymorphisms
(SNPs) or mutations associated with ESCC (26–29);
however, the majority of these do not have biological functions.
Thus, increasing attention is being focused on the identification
of oncogenes or tumor suppressors. The present study investigated
the transcription factor ATF3 and observed that its expression was
reduced in ESCC, and that increased expression of ATF3 resulted in
tumor inhibition, characterized by suppression of cell
proliferation, motility and migration. In addition, it was noted
that ATF3 also negatively regulated the oncogene ID1, and repressed
the expression of cyclin D1, STAT3 and TWIST.
Controversial roles of ATF3 have been reported in
various types of cancer in which ATF3 exhibits oncogenic or
suppressive functions in cancer formation and progression.
Consistent with a recent study of ESCC (21), the results of the current study
demonstrated that ATF3 expression was lower in the ESCC tissues
compared with the adjacent non-tumor tissues, and the expression
levels negatively correlated with cancer differentiation, thus
indicating the tumor suppressive functions of ATF3 in ESCC. By
contrast, expression levels of the oncogene ID1 were higher in the
ESCC tissues compared with the adjacent non-tumor tissues. ATF3 and
ID1 expression demonstrated a significant inverse correlation in
the ESCC tissues. In vitro experiments further indicated
that increased expression of ATF3 repressed ID1 expression at the
protein and mRNA level in ESCC cells, and that there was a negative
regulatory interaction between ATF3 and ID1, supported by results
from co-immunoprecipitation assays. The present study therefore
provides additional evidence that ATF3 inhibits ID1 expression
through a protein-protein interaction.
Tumor formation is understood to result from
uncontrollable cell growth, which is associated with overexpression
of cell cycle regulators (30),
including cyclin dependent kinases, cyclins (e.g. cyclin D1) and
cyclin dependent kinase inhibitors. Numerous studies have reported
that cyclin D1 is overexpressed in cancer tissues and inhibition of
cyclin D1 expression in vitro leads to cell proliferation
inhibition (31,32). In the present study, it was
demonstrated that increased expression of ATF3 repressed cyclin D1
expression in the ESCC cells. Furthermore, STAT3 is known to serve
a crucial role in carcinogenesis, particularly the activated form
of STAT3, p-STAT3 (33,34). The current study observed that
increased ATF3 expression was able to suppress the levels of
p-STAT3 in the ESCC cell lines, and the downregulation of cyclin D1
and p-STAT3 partially led to inhibition of ESCC cell proliferation
in vitro.
Cancer-associated mortality is primarily caused by
metastasis, and the latter is linked to the upregulation of TWIST
and downregulation of E-cadherin (35–40). A
number of studies have reported that loss of E-cadherin expression
increases cancer metastasis by separating cancer cells from
another, thus activating specific downstream signal transduction
pathways resulting in epithelial-mesenchymal transition (41–45).
Perturbation of E-cadherin-mediated cell adhesion is implicated in
the progression of tumors, poor prognosis and metastasis (35–40). An
increasing body of evidence indicates the importance of E-cadherin
in cancer metastasis and development, and the possible regulation
of E-cadherin by glycogen synthase kinase 3β, TWIST/Snail and
Akt/protein kinase B (41–43). Maintaining the expression of
E-cadherin may prevent tumor invasion and metastasis and restore
epithelial morphology (44,45). In the present study, it was observed
that increasing ATF3 expression upregulated E-cadherin expression
in the ESCC cells and downregulated the expression of Twist,
resulting in the inhibition of ESCC cell migration in
vitro.
In conclusion, the current study demonstrated that
ATF3 expression was decreased and ID1 expression was overexpressed
in ESCC tissues; however, increasing ATF3 expression led to
inhibition of cell proliferation, migration and motility in
vitro, which was associated with the upregulation of E-cadherin
and the downregulation of cyclin D1 and Twist, and most importantly
highlighted the inverse regulatory interaction between ATF3 and
ID1. These results provide additional evidence of the tumor
suppressive features of ATF3 and demonstrate the novel mechanism of
ATF3-mediated inhibition of cancer metastasis in esophageal
cancer.
Acknowledgements
The current study was supported in part by the
Provincial Natural Science Foundation (grant no. 122300410401) from
the Science and Technology Department of Henan Province, China, and
the US Chinese Anti-Cancer Association, CA, USA (grant no.
USCACA-TIGM-1).
Glossary
Abbreviations
Abbreviations:
|
ESCC
|
esophageal squamous cell carcinoma
|
|
ATF3
|
activating transcription factor 3
|
|
ID1
|
inhibitor of DNA binding 1
|
|
STAT3
|
signaling transducer and activator of
transcription 3
|
References
|
1
|
Thompson MR, Xu D and Williams BR: ATF3
transcription factor and its emerging roles in immunity and cancer.
J Mol Med (Berl). 87:1053–1060. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yin X, Dewille JW and Hai T: A potential
dichotomous role of ATF3, an adaptive-response gene, in cancer
development. Oncogene. 27:2118–2127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pelzer AE, Bektic J, Haag P, Berger AP,
Pycha A, Schäfer G, Rogatsch H, Horninger W, Bartsch G and Klocker
H: The expression of transcription factor activating transcription
factor 3 in the human prostate and its regulation by androgen in
prostate cancer. J Urol. 175:1517–1522. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Janz M, Hummel M, Truss M, Wollert-Wulf B,
Mathas S, Jöhrens K, Hagemeier C, Bommert K, Stein H, Dörken B and
Bargou RC: Classical Hodgkin lymphoma is characterized by high
constitutive expression of activating transcription factor 3
(ATF3), which promotes viability of Hodgkin/Reed-Sternberg cells.
Blood. 107:2536–2539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ling MT, Lau TC, Zhou C, Chua CW, Kwok WK,
Wang Q, Wang X and Wong YC: Overexpression of Id-1 in prostate
cancer cells promotes angiogenesis through the activation of
vascular endothelial growth factor (VEGF). Carcinogenesis.
26:1668–1676. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bandyopadhyay S, Wang Y, Zhan R, Pai SK,
Watabe M, Iiizumi M, Furuta E, Mohinta S, Liu W, Hirota S, et al:
The tumor metastasis suppressor gene Drg-1 down-regulates the
expression of activating transcription factor 3 in prostate cancer.
Cancer Res. 66:11983–11990. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maytin EV, Ubeda M, Lin JC and Habener JF:
Stress-inducible transcription factor CHOP/gadd153 induces
apoptosis in mammalian cells via p38 kinase-dependent and
-independent mechanisms. Exp Cell Res. 267:193–204. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan X, Yu L, Li J, Xie G, Rong T, Zhang
L, Chen J, Meng Q, Irving AT, Wang D, et al: ATF3 suppresses
metastasis of bladder cancer by regulating gelsolin-mediated
remodeling of the actin cytoskeleton. Cancer Res. 73:3625–3637.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jan YH, Tsai HY, Yang CJ, Huang MS, Yang
YF, Lai TC, Lee CH, Jeng YM, Huang CY, Su JL, et al: Adenylate
kinase-4 is a marker of poor clinical outcomes that promotes
metastasis of lung cancer by downregulating the transcription
factor ATF3. Cancer Res. 72:5119–5129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei S, Wang H, Lu C, Malmut S, Zhang J,
Ren S, Yu G, Wang W, Tang DD and Yan C: The activating
transcription factor 3 protein suppresses the oncogenic function of
mutant p53 proteins. J Biol Chem. 289:8947–8959. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kang Y, Chen CR and Massague J: A
self-enabling TGFbeta response coupled to stress signaling: Smad
engages stress response factor ATF3 for Id1 repression in
epithelial cells. Mol Cell. 11:915–926. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bottone FG Jr and Alston-Mills B: The
dietary compounds resveratrol and genistein induce activating
transcription factor 3 while suppressing inhibitor of DNA
binding/differentiation-1. J Med Food. 14:584–593. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Perk J, Iavarone A and Benezra R: Id
family of helix-loop-helix proteins in cancer. Nat Rev Cancer.
5:603–614. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Benezra R, Davis RL, Lockshon D, Turner DL
and Weintraub H: The protein Id: A negative regulator of
helix-loop-helix DNA binding proteins. Cell. 61:49–59. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patel D, Morton DJ, Carey J, Havrda MC and
Chaudhary J: Inhibitor of differentiation 4 (ID4): From development
to cancer. Biochim Biophys Acta. 1855:92–103. 2014.PubMed/NCBI
|
|
16
|
Castanon E, Bosch-Barrera J, López I,
Collado V, Moreno M, López-Picazo JM, Arbea L, Lozano MD, Calvo A
and Gil-Bazo I: Id1 and Id3 co-expression correlates with clinical
outcome in stage III-N2 non-small cell lung cancer patients treated
with definitive chemoradiotherapy. J Transl Med. 11:132013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang R, Hirsch P, Fava F, Lapusan S,
Marzac C, Teyssandier I, Pardo J, Marie JP and Legrand O: High Id1
expression is associated with poor prognosis in 237 patients with
acute myeloid leukemia. Blood. 114:2993–3000. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schoppmann SF, Schindl M, Bayer G, Aumayr
K, Dienes J, Horvat R, Rudas M, Gnant M, Jakesz R and Birner P:
Overexpression of Id-1 is associated with poor clinical outcome in
node negative breast cancer. Int J Cancer. 104:677–682. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kashiwakura Y, Ochiai K, Watanabe M,
Abarzua F, Sakaguchi M, Takaoka M, Tanimoto R, Nasu Y, Huh NH and
Kumon H: Down-regulation of inhibition of differentiation-1 via
activation of activating transcription factor 3 and Smad regulates
REIC/Dickkopf-3-induced apoptosis. Cancer Res. 68:8333–8341. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tan H, Zhang H, Xie J, Chen B, Wen C, Guo
X, Zhao Q, Wu Z, Shen J, Wu J, et al: A novel staging model to
classify oesophageal squamous cell carcinoma patients in China. Br
J Cancer. 110:2109–2115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie JJ, Xie YM, Chen B, Pan F, Guo JC,
Zhao Q, Shen JH, Wu ZY, Wu JY, Xu LY and Li EM: ATF3 functions as a
novel tumor suppressor with prognostic significance in esophageal
squamous cell carcinoma. Oncotarget. 5:8569–8582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li T, Yang W, Li M, Byun DS, Tong C,
Nasser S, Zhuang M, Arango D, Mariadason JM and Augenlicht LH:
Expression of selenium-binding protein 1 characterizes intestinal
cell maturation and predicts survival for patients with colorectal
cancer. Mol Nutr Food Res. 52:1289–1299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bi X, Pohl NM, Qian Z, Yang GR, Gou Y,
Guzman G, Kajdacsy-Balla A, et al: Decorin-mediated inhibition of
colorectal cancer growth and migration is associated with
E-cadherin in vitro and in mice. Carcinogenesis. 33:326–330. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang LD, Zhou FY, Li XM, Sun LD, Song X,
Jin Y, Li JM, Kong GQ, Qi H, Cui J, et al: Genome-wide association
study of esophageal squamous cell carcinoma in Chinese subjects
identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet.
42:759–763. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Abnet CC, Freedman ND, Hu N, Wang Z, Yu K,
Shu XO, Yuan JM, Zheng W, Dawsey SM, Dong LM, et al: A shared
susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma
and esophageal squamous cell carcinoma. Nat Genet. 42:764–767.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y,
Liu Z, Zhan Q, Liu Y, Yu D, et al: Genome-wide association study
identifies three new susceptibility loci for esophageal
squamous-cell carcinoma in Chinese populations. Nat Genet.
43:679–684. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu C, Wang Z, Song X, Feng XS, Abnet CC,
He J, Hu N, Zuo XB, Tan W, Zhan Q, et al: Joint analysis of three
genome-wide association studies of esophageal squamous cell
carcinoma in Chinese populations. Nat Genet. 46:1001–1006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shtutman M, Zhurinsky J, Simcha I,
Albanese C, et al: The cyclin D1 gene is a target of the
beta-catenin/LEF-1 pathway. PNAS U.S.A. 96:5522–5527. 1999.
View Article : Google Scholar
|
|
31
|
Wu S, Bao Y, Ma D, Zi Y, et al: Sodium
selenite inhibits leukemia HL-60 cell proliferation and induces
cell apoptosis by enhancing the phosphorylation of JNK1 and
increasing the expression of p21 and p27. Int J Mol Med.
34:1175–1179. 2014.PubMed/NCBI
|
|
32
|
Okumura H, Uchikado Y, Setoyama T,
Matsumoto M, Owaki T, Ishigami S and Natsugoe S: Biomarkers for
predicting the response of esophageal squamous cell carcinoma to
neoadjuvant chemoradiation therapy. Surg Today. 44:421–428. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kamran MZ, Patil P and Gude RP: Role of
STAT3 in cancer metastasis and translational advances. Biomed Res
Int. 2013:4218212013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Quante M, Varga J, Wang TC and Greten FR:
The gastrointestinal tumor microenvironment. Gastroenterology.
145:63–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Doki Y, Shiozaki H, Tahara H, Inoue M, Oka
H, Iihara K, Kadowaki T, Takeichi M and Mori T: Correlation between
E-cadherin expression and invasiveness in vitro in a human
esophageal cancer cell line. Cancer Res. 53:3421–3426.
1993.PubMed/NCBI
|
|
36
|
Oka H, Shiozaki H, Kobayashi K, Inoue M,
Tahara H, Kobayashi T, Takatsuka Y, Matsuyoshi N, Hirano S,
Takeichi M, et al: Expression of E-cadherin cell adhesion molecules
in human breast cancer tissues and its relationship to metastasis.
Cancer Res. 53:1696–1701. 1993.PubMed/NCBI
|
|
37
|
Umbas R, Isaacs WB, Bringuier PP,
Schaafsma HE, Karthaus HF, Oosterhof GO, Debruyne FM and Schalken
JA: Decreased E-cadherin expression is associated with poor
prognosis in patients with prostate cancer. Cancer Res.
54:3929–3933. 1994.PubMed/NCBI
|
|
38
|
Derksen PW, Liu X, Saridin F, van der
Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink
J, Krimpenfort P, et al: Somatic inactivation of E-cadherin and p53
in mice leads to metastatic lobular mammary carcinoma through
induction of anoikis resistance and angiogenesis. Cancer Cell.
10:437–449. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Frixen UH, Behrens J, Sachs M, Eberle G,
Voss B, et al: E-cadherin-mediated cell-cell adhesion prevents
invasiveness of human carcinoma cells. J Cell Biol. 113:173–185.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Onder TT, Gupta PB, Mani SA, Yang J,
Lander ES and Weinberg RA: Loss of E-cadherin promotes metastasis
via multiple downstream transcriptional pathways. Cancer Res.
68:3645–3654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Baranwal S and Alahari SK: Molecular
mechanisms controlling E-cadherin expression in breast cancer.
Biochem Biophys Res Commun. 384:6–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang CC and Wolf DA: Inflamed snail speeds
metastasis. Cancer Cell. 15:355–357. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schmalhofer O, Brabletz S and Brabletz T:
E-cadherin, beta-catenin, and ZEB1 in malignant progression of
cancer. Cancer Metastasis Rev. 28:151–166. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nelson WJ and Nusse R: Convergence of Wnt,
beta-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
View Article : Google Scholar : PubMed/NCBI
|