Introduction
Renal cell carcinoma (RCC) represents the most
common malignancy of the adult kidney and comprises 2–3% of all
malignant tumors in adults (1). In
2014, there were 63,920 novel cases of kidney cancer diagnosed in
the USA (2). RCC accounts for ~90% of
all kidney malignancies, with a rising incidence over the past
decade (3). RCC is an epithelial
neoplasm, since it originates from the transformation of cells that
constitute the epithelium of the proximal convoluted tubule
(4). RCC includes several
histological subtypes that possess distinct biological behaviors
and prognoses. The principal histological subtypes are as follows:
Clear cell RCC (75–80% of RCC); papillary or chromophile RCC
(10–15%); and chromophobe RCC (4–6%) (4). Although the majority of patients with
early-stage RCC may be cured surgically, 10–20% of patients present
with metastasis at the time of diagnosis (5–7). In
addition, ~50% of RCC patients that have undergone curative surgery
may be expected to develop a recurrence with distant metastasis,
and the prognosis of RCC patients with metastatic or recurrent
disease is poor, with a 5-year survival rate of <20% (8). The most common sites of metastatic
spread in RCC are lung, bone, adrenal gland, liver and brain, and
more than one organ is often involved (9). Therefore, the molecular mechanisms of
cancer cell dissemination from the primary tumor are important when
considering treatment and observational strategies for RCC
patients.
MicroRNAs (miRs) are 21–25 nucleotide
single-stranded, non-coding RNA molecules that exert their
functions by binding to the 3′-untranslated regions (UTRs) of their
corresponding mRNA targets, which leads to target mRNA cleavage or
translational repression (10). It
has been widely accepted that miRs are important in numerous
biological processes, including differentiation, proliferation,
apoptosis, cell cycle, migration and invasion (11). Research has revealed that a single miR
targets hundreds of mRNAs, and ~50% of miR genes are located in
cancer-associated chromosomal regions (12). In addition, mature miRs regulate the
expression of ~10–30% of all human genes (13). miRs function as either tumor
suppressors or oncogenes, depending on the genes that they target
(14); tumor suppressive miRs are
usually downregulated, while oncogenic miRs are upregulated in
cancer (15). Recent evidence
indicates that the aberrant regulation of miRs are important in RCC
pathogenesis (16). Therefore, miRs
are being investigated as potential biomarkers for diagnosis and
prognosis of human malignancies, due to their tissue and
disease-specific expression and regulatory functions (17).
miR-429 has been frequently reported to be
downregulated in various tumors, including RCC (18), nasopharyngeal carcinoma (19), Ehrlich ascites tumor cells (20), gastric cancer (21), non-small cell lung cancer (22) and endometrial endometrioid carcinoma
(23). However, the function of
miR-429 has yet to be elucidated in RCC. The aim of the present
study was to investigate the effect of miR-429 on RCC and to
elucidate its underlying mechanisms.
Materials and methods
Cell culture and transfection
Human RCC 786-O and A498 cell lines were purchased
from the Shanghai Institute of Cell Biology, Chinese Academy of
Science (Shanghai, China). The cells were cultured in RPMI-1640
medium (Gibco®; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) supplemented with 10% heat-inactivated fetal calf serum
(FCS; Gibco®), 100 U/ml penicillin and 100 mg/l
streptomycin. Cultures were maintained at 37°C in a humidified
atmosphere with 5% CO2.
Mature miR-429 mimics (UAAUACUGUCUGGUAAAACCGU),
scrambled control (NC; UUCUCCGAACGUGUCACGUTT) and luciferase
reporter plasmid were purchased from Shanghai GenePharma Co., Ltd.
(Shanghai, China). Transient transfection of miR-429 mimics and/or
plasmid were performed using Lipofectamine® 2000
(Invitrogen™; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol.
Cell viability assay
A MTT assay was used to estimate the proliferation
ability of the cells. Cells transfected with miR-429 mimics or NC
were seeded in 96-well plates at a density of 3,000 cells/well for
24, 48, 72, 96, 120 and 144 h. Briefly, the cells were incubated
with 20 ml MTT (5 mg/ml) for 4 h at 37°C. The plates were agitated
and 200 µl dimethyl sulfoxide was added to solubilize the crystals
for 20 min following the removal of the culture supernatant.
Absorbance was measured at 490 nm using an automatic multi-well
spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The suppression rate was calculated using the following formula:
Suppression rate = [1 - optical density (ODmiR-429 /
ODNC)] × 100. All the experiments were performed in
triplicate.
Cell migration and invasion assay
Cell migration and invasion assays were evaluated
using Transwell chambers (Corning Incorporated, Corning, NY, USA).
The upper and lower culture compartments were separated by
polycarbonate filters with a 8 mm pore size. Prior to the invasion
assays, the filters of the Transwell chambers were coated with 30
µg Matrigel (BD Biosciences, San Jose, CA, USA) to form a
reconstituted basement membrane. Transfected cells (miR-429 mimics
and NC) in the log phase were treated with trypsin/EDTA solution,
washed once with serum-containing RPMI-1640 medium, centrifuged (at
200 × g for 10 min) and re-suspended as single-cell solutions. A
total of 1×105 cells in 0.2 ml serum-free RPMI-1640
medium were seeded into the Transwell chambers. The lower chamber
contained RPMI −1640 supplemented with 20% FCS to stimulate
invasion. The cells were incubated for 12 h for the migration assay
and 24 h for the invasion assay. Non-migrating and -invading cells
on the top of the membrane were removed by scraping. Cells that
migrated to the bottom surface of the insert were fixed with 100%
methanol, stained with 0.5% crystal violet, and subjected to
microscopic inspection (magnification, ×200). Values for invasion
and migration were obtained by counting five fields per membrane,
and represent the average of three independent experiments.
Western blotting
Western blotting was performed as described
previously (24). Primary antibodies
used in the present study were rabbit anti-human specificity
protein 1 (SP1) monoclonal antibody (1:1,000 dilution; catalog no.
9389; Cell Signaling Technology, Inc., Danvers, MA, USA) and rabbit
anti-human anti-β-actin monoclonal antibody (1:1,000 dilution;
catalog no. AP0060; Bioworld Technology, Inc., Louis Park, MN,
USA). Total protein was extracted using ice-cold
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Jiangsu, Haimen, China). Equal amounts of protein
were subjected to 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis and transferred to polyvinylidene difluoride
membranes (Bio-Rad Laboratories, Inc.). Following blocking with 5%
non-fat milk in Tris-buffered saline with 0.1% Tween 20, the
membranes were incubated overnight at 4°C with the appropriate
primary antibody. Following washing, the blots were incubated with
goat anti-rabbit horseradish peroxidase-conjugated secondary
antibody (1:2,000 dilution; catalog no. 7074; Cell Signaling
Technology, Inc.) at room temperature for 1 h. The intensity of
each blot was read and analyzed with AlphaEaseFC™ software (version
4.0.1; Cell Biosciences, Palo Alto, CA, USA). β-actin was used as a
loading control.
Luciferase assay
Reporter plasmids (pmirGLO-SP1-3′UTR wild-type and
pmirGLO-SP1-3′UTR mutant) were synthesized and purified by
GenePharma. Cells were plated in a 12-well plate at ~90% confluence
and transfected with reporter plasmid, miR-429 mimics or NC using
Lipofectamine 2000. Each sample was also cotransfected with 0.05 µg
pRL-CMV plasmid expressing Renilla Luciferase (Promega Corporation,
Madison, WI, USA), as an internal control for transfection
efficiency. Subsequent to 24 h, luciferase activity was measured
using the Dual-Luciferase® Reporter Assay System
(Promega Corporation), according to the manufacturer's protocol.
Firefly luciferase activities and Renilla luciferase activities
were measured with a luminometer (Tecan Group, Ltd, Männedorf,
Switzerland). Each assay was replicated three times.
Statistical analysis
Data are presented as the mean ± standard deviation,
and compared using Student's t-test in Stata version 10.0 software
(StataCorp LP, College Station, TX, USA). Double-tailed P<0.05
was considered to indicate a statistically significant
difference.
Results
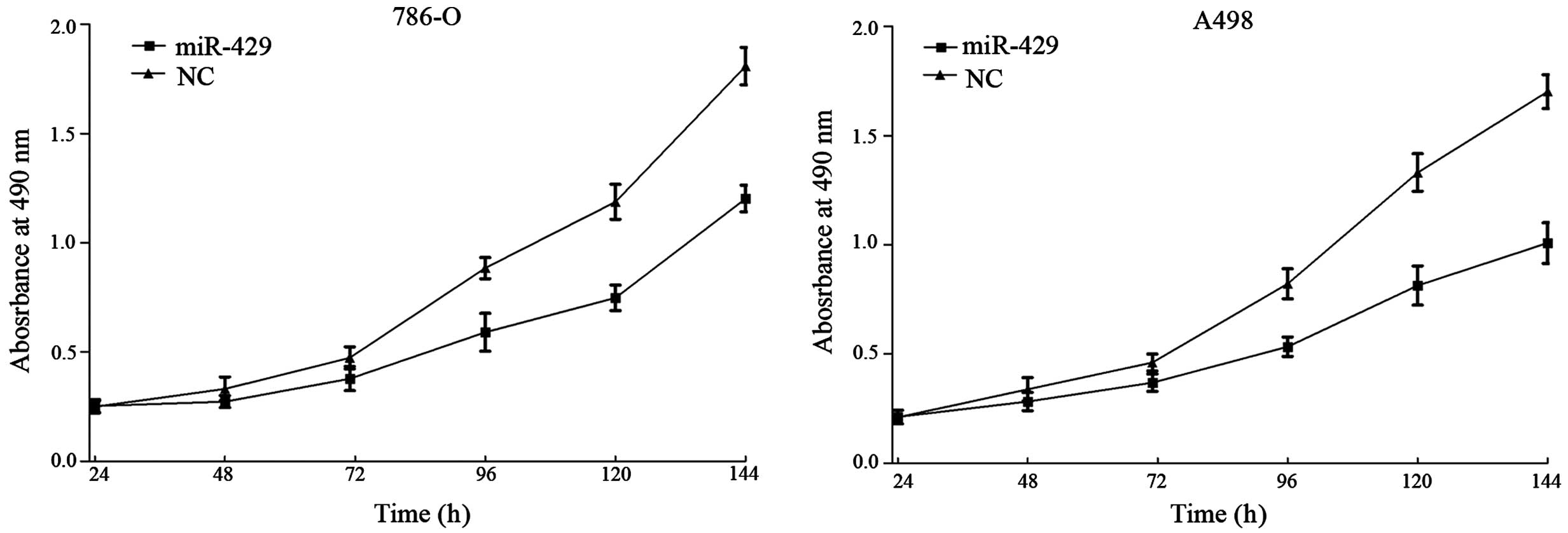
miR-429 suppresses cell proliferation
in RCC cell lines
MTT assays were employed to detect the proliferation
of 786-O and A498 cell lines following transfection with miR-429 or
NC. As shown in Fig. 1,
overexpression of miR-429 in 786-O and A498 cells inhibited cell
proliferation. Compared with the NC-transfected cells, the
suppression rate of miR-429 reached 33.50±2.90% in 786-O cells
(P=0.010) and 43.31±4.20% in A498 cells (P=0.002) following 144 h
of treatment.
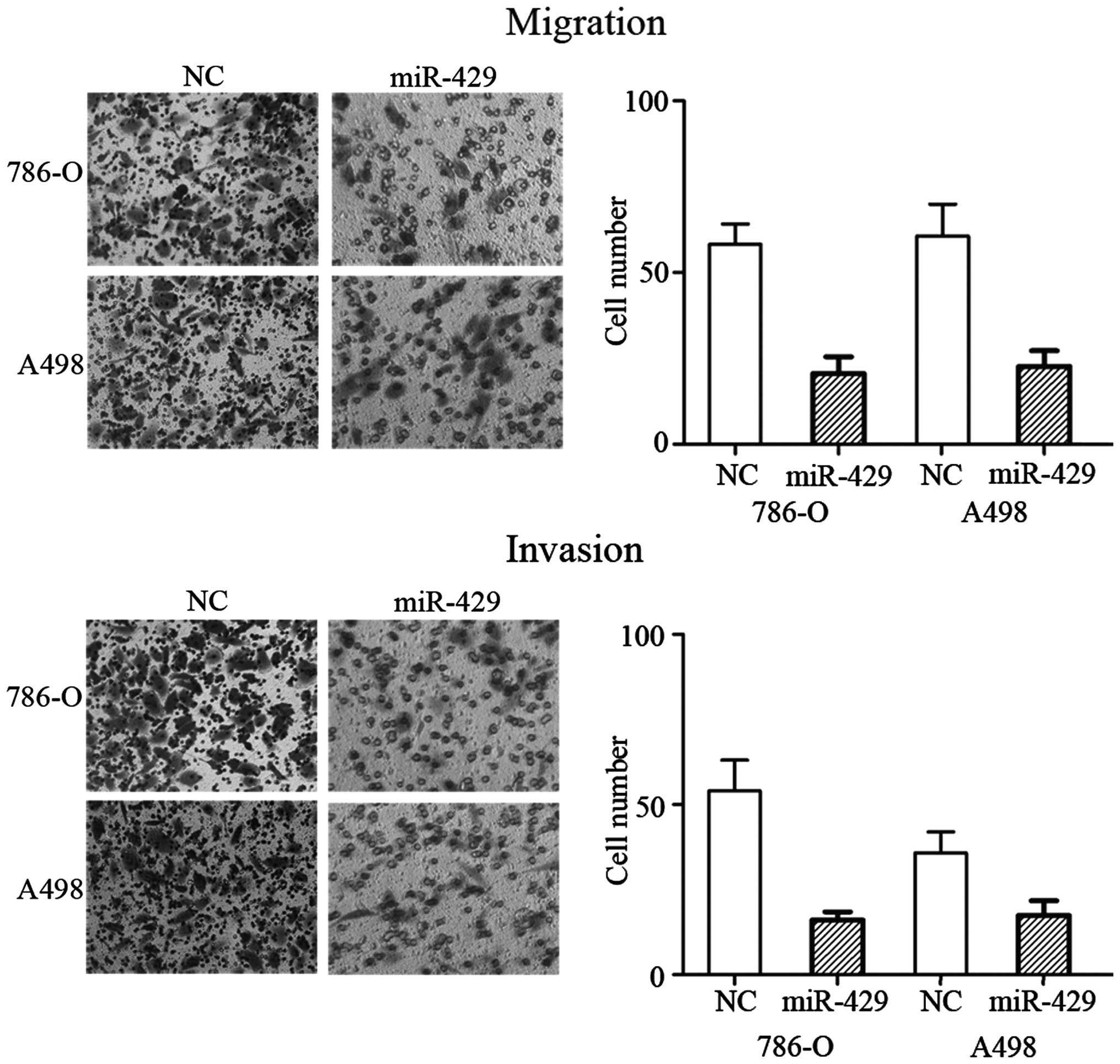
miR-429 inhibits cell migration and
invasion in RCC cell lines
To determine whether miR-429 regulates human RCC
cell migration and invasion, migration and invasion assays were
performed on miR-429 mimic- and NC-transfected RCC 786-O and A498
cell lines. As expected, overexpression of miR-429 significantly
decreased the migration (P=0.024 for 786-O cells and P=0.029 for
A498 cells) and invasion (P=0.020 for 786-O cells and P=0.039 for
A498 cells) capability of the 786-O and A498 cells (Fig. 2). These observations indicated that
miR-429 was a negative regulator of RCC migration and invasion.
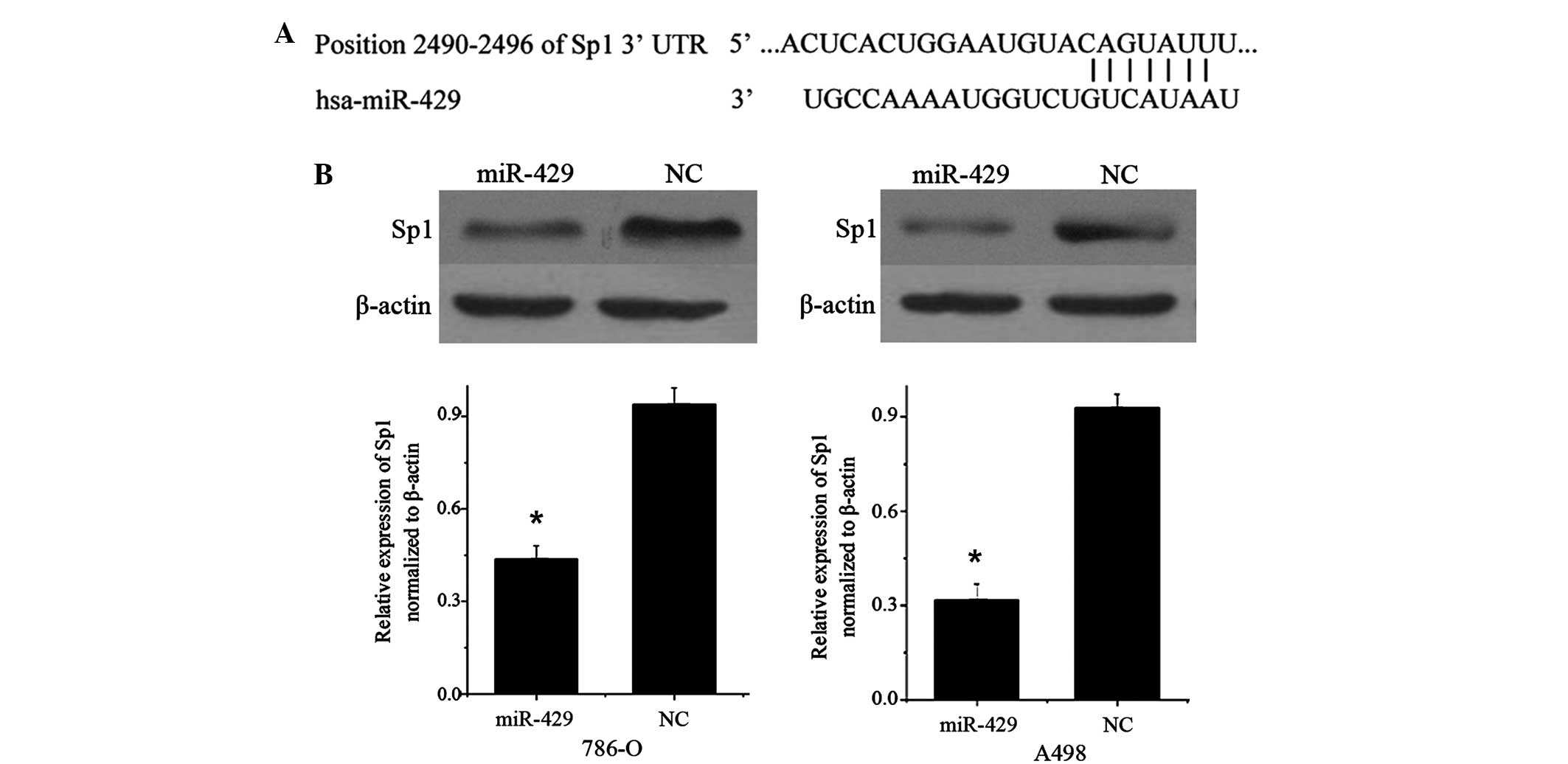
miR-429 suppresses the expression of
Sp1 in RCC cell lines
TargetScan (version 5.2; www.targetscan.org) predicted that Sp1 was a direct
target gene of miR-429, and revealed that Sp1 mRNA contained a
miR-429 7-nucleotide seed match at position 2490–2496 of the Sp1
3′-UTR (Fig. 3A).
Therefore, the present study performed western blot
analysis to investigate whether Sp1 protein level was decreased
following overexpression of miR-429. As shown in Fig. 3B, Sp1 was significantly decreased in
786-O (P=0.018) and A498 cell lines (P=0.012) 72 h subsequent to
transfection of miR-429 compared with cells transfected with NC.
These data reveal that Sp1 is a direct functional target of
miR-429.
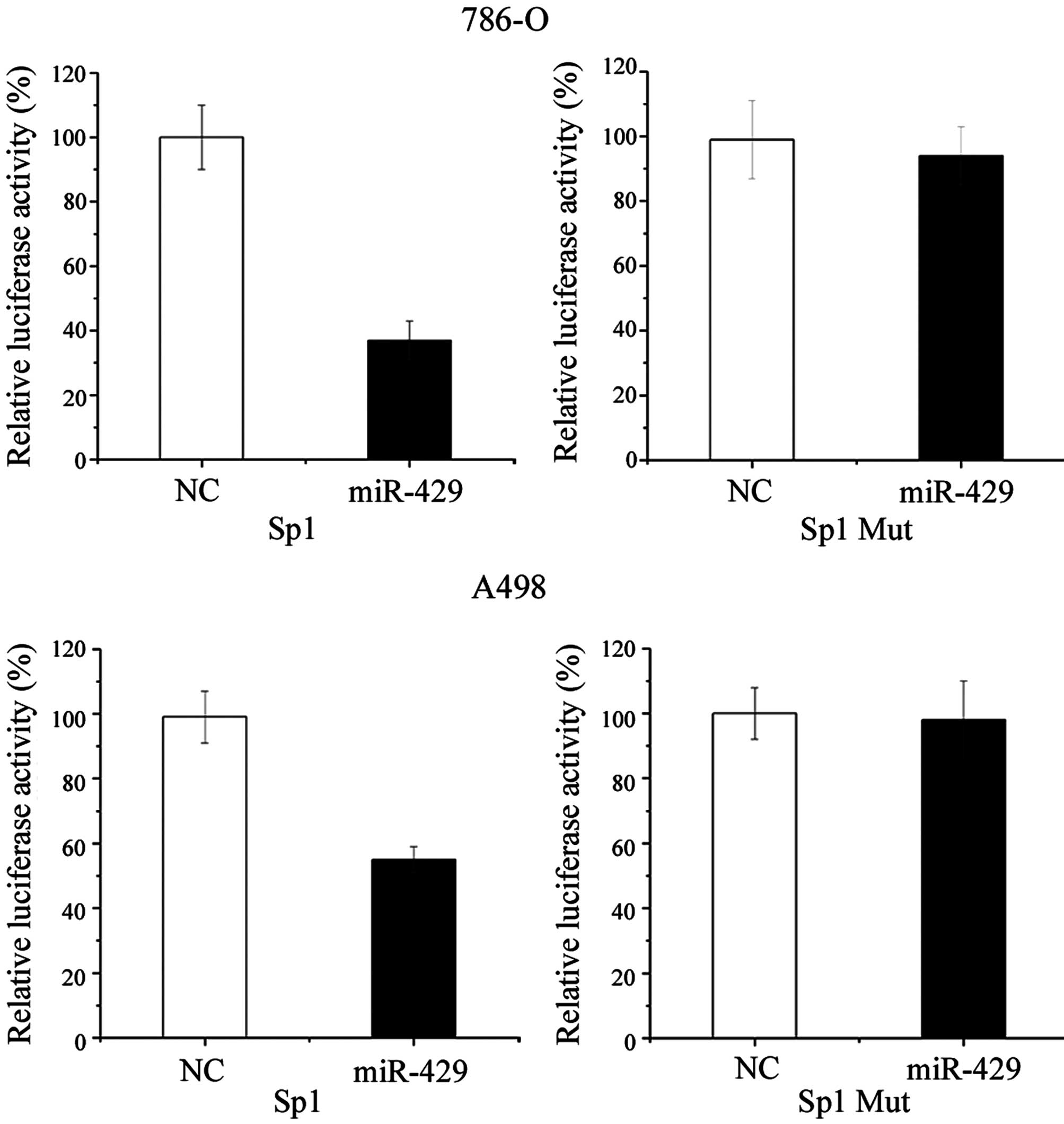
Sp1 is a direct target of miR-429
To further confirm that Sp1 is a direct target for
miR-429, a luciferase reporter assay was performed in RCC 786-O and
A498 cell lines. As shown by Fig. 4,
transfection of cells with miR-429 mimic significantly decreased
the Sp1 3′-UTR luciferase reporter activity compared with cells
transfected with NC. Overexpression of miR-429 suppressed Sp1
3′-UTR-luciferase activity by 63% in 786-O cells (P=0.020) and 44%
in A498 cells (P=0.034). In addition, this effect was abolished
when the nucleotides in the seed binding sites of the Sp1 3′-UTR
were mutated (Fig. 4).
The present results suggest that Sp1 is a target
gene of miR-429.
Discussion
miR-429 belongs to a small miR family, which
includes miR-200c, miR-141, miR-200b and miR-200a (25), and is located on chromosome 1.
Downregulation of miR-429 may be important in tumor progression,
and previous studies have revealed that miR-429 is frequently
downregulated in various tumors, including RCC (18), nasopharyngeal carcinoma (19), Ehrlich ascites tumor cells (20), gastric cancer (21), non-small cell lung cancer (22) and endometrial endometrioid carcinoma
(23). However, miR-429 has been
demonstrated to be upregulated in bladder cancer, and an increased
expression was correlated with poor prognosis in serous ovarian
carcinoma patients (26,27). Therefore, miR-429 appears to be
involved in carcinogenesis and cancer progression, and plays
various roles depending on the type of cancer.
In gastric cancer, miR-429 inhibits the
proliferation, migration and invasion of tumor cells. In addition,
the expression levels of miR-429 in the tissues of gastric cancer
patients with lymph node metastasis were significantly lower
compared with patients without lymph node metastasis (28). In colorectal cancer, miR-429 was
revealed to be significantly downregulated in the majority of tumor
tissues investigated, particularly tissue from stages II and III,
relative to adjacent or distant normal mucosa. Survival analysis
also indicated a low expression of miR-429 is significantly
associated with poor prognosis (29).
Upregulation of miR-429 suppressed cell growth, invasion and the
expression of epithelial-mesenchymal transition-association marker
genes, including E-cadherin, catenin (CTNN) A1, CTNNB1,
transferring N-terminal half-molecule, cluster of differentiation
44, matrix metallopeptidase 2, vimentin, Slug, Snail and zinc
finger E-box binding homeobox (ZEB) 2 (18). These results suggest that
downregulation of miR-429 in tumour cells may play a role in the
development of cancer by enhancing cell proliferation and promoting
cell migration and invasion.
Identification of miR-429 target genes is critical
for understanding the role of miR-429 in tumorigenesis, and is
important for defining novel therapeutic targets. Previous studies
have demonstrated that miR-429 regulates oncogenic expression in
human cells, including c-myc (28),
onecut 2 (18), ZEB1 (30), v-crk avian sarcoma virus CT10 oncogene
homolog-like (31) and B-cell
lymphoma 2 (32). Therefore,
upregulation of miR-429 or providing analogous pharmaceutical
compounds exogenously may be effective cancer therapy for RCC,
leading to the regulation of these oncogenic transcripts. The
present results suggest multiple inhibitory effects of miR-429 in
human RCC 786-O and A498 cell lines, including cell proliferation
and invasion suppression, via the downregulation of the expression
of Sp1. The present findings suggest that miR-429 may be used for
the development of novel molecular markers and therapeutics for
RCC.
Sp1 is an ubiquitously expressed transcription
factor, and was the first transcription factor to be cloned from
mammalian cells in 1983 (33). It
maps to 12q13.1 and encodes a protein of 785 amino acids (34). Sp1 recognizes GC-rich regions and
binds to DNA through three C2H2-type zinc fingers in the C-terminal
domain (35,36). Each zinc finger of Sp1 recognizes
three bases in one strand and a single base in the complementary
strand, constituting a consensus binding sequence of 5′-(G/T)
GGGCGG (G/A) (G/A) (C/T)-3′ (37).
Originally, Sp1 was considered to be a general transcription factor
required for transcription of a large number of ‘housekeeping
genes’ (38). However, recently a
study has indicated that many of these housekeeping genes are
crucial in tumorigenesis and cancer progression (39). Consequently, Sp1 has been demonstrated
to be important in tumor progression, including cell proliferation,
angiogenesis, differentiation, apoptosis, migration and invasion,
revealing it as an ideal target for cancer treatment (40).
Sp1 has been identified to be upregulated in various
types of human cancer, including breast carcinoma (41), thyroid cancer (42), hepatocellular carcinoma (43), pancreatic cancer (44), colorectal cancer (45), gastric cancer (46) and lung cancer (47). Sp1 expression is tightly regulated
throughout the various stages of tumorigenesis, which affects
cancer progression (48). Previous
studies concerning Sp1 expression in tumorigenesis suggest that
increased transcription is the most important mechanism for Sp1
accumulation during cancer formation (49,50). Sp1
contributes to cancer progression and is regulated by many miRs. In
non-small cell lung cancer cells, miR-335 and miR-27b suppressed
the proliferation, invasion and induced apoptosis in cells by
targeting Sp1 (51,52). In prostate cancer, Mao et al
(53) demonstrated that miR-330
suppressed cell motility by targeting Sp1. In squamous cervical
cancer, Wang et al (54)
revealed that miR-375 inhibited cell migration and invasion by
targeting Sp1. The present study revealed that miR-429 suppressed
RCC cell viability, migration and invasion via the downregulation
of Sp1. Therefore, Sp1 may be investigated as a predictive marker
for the early detection of tumor recurrence, and be used in
therapeutics to block RCC cells from becoming invasive.
In summary, the present study is, to the best of our
knowledge, the first study to demonstrate that miR-429 contributes
to cell viability, migration and invasion in RCC. The
identification of candidate target genes of miR-429, such as Sp1,
may provide an understanding of the potential carcinogenic
mechanisms in RCC. These findings have therapeutic implications and
may be used for novel treatment strategies for RCC.
References
|
1
|
Escudier B: Emerging immunotherapies for
renal cell carcinoma. Ann Oncol. 23(Suppl 8): viii35–viii40. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu S, Wu H, Nie H, Yue L, Jiang H, Xiao S
and Li Y: AIF downregulation and its interaction with STK3 in renal
cell carcinoma. PLoS One. 9:e1008242014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Srigley JR, Delahunt B, Eble JN, Egevad L,
Epstein JI, Grignon D, Hes O, Moch H, Montironi R, Tickoo SK, et
al: The international society of urological pathology (ISUP)
vancouver classification of renal neoplasia. Am J Surg Pathol.
37:1469–1489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang YQ and Chen J: Predictive role of
vascular endothelial growth factor polymorphisms in the survival of
renal cell carcinoma patients. Genet Mol Res. 13:5011–5017. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Flanigan RC, Campbell SC, Clark JI and
Picken MM: Metastatic renal cell carcinoma. Curr Treat Options
Oncol. 4:385–390. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Athar U and Gentile TC: Treatment options
for metastatic renal cell carcinoma: A review. Can J Urol.
15:3954–3966. 2008.PubMed/NCBI
|
|
8
|
Irani J: Sunitinib versus interferon-alpha
in metastatic renal-cell carcinoma. Prog Urol. 17:9962007.(In
French). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Toma MI, Erdmann K, Diezel M, Meinhardt M,
Zastrow S, Fuessel S, Wirth MP and Baretton GB: Lack of ephrin
receptor A1 is a favorable independent prognostic factor in clear
cell renal cell carcinoma. PLoS One. 9:e1022622014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lai EC: Micro RNAs are complementary to 3′
UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miao J, Wu S, Peng Z, Tania M and Zhang C:
MicroRNAs in osteosarcoma: Diagnostic and therapeutic aspects.
Tumour Biol. 34:2093–2098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma XP, Zhang T, Peng B, Yu L and Jiang de
K: Association between microRNA polymorphisms and cancer risk based
on the findings of 66 case-control studies. PLoS One. 8:e795842013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Berezikov E, Guryev V, van de Belt J,
Wienholds E, Plasterk RH and Cuppen E: Phylogenetic shadowing and
computational identification of human microRNA genes. Cell.
120:21–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang
Z, Wang X, Lin Y, Mao Y, et al: Downregulation of microRNA-182-5p
contributes to renal cell carcinoma proliferation via activating
the AKT/FOXO3a signaling pathway. Mol Cancer. 13:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartels CL and Tsongalis GJ: MicroRNAs:
Novel biomarkers for human cancer. Ann Biol Clin (Paris).
68:263–272. 2010.(In French). PubMed/NCBI
|
|
18
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: MiR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bartis D, Mise N, Mahida RY, Eickelberg O
and Thickett DR: Epithelial-mesenchymal transition in lung
development and disease: Does it exist and is it important? Thorax.
69:760–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Said NA and Williams ED: Growth factors in
induction of epithelial-mesenchymal transition and metastasis.
Cells Tissues Organs. 193:85–97. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu W, He J, Chen D, Zhang B, Xu L, Ma H,
Liu X, Zhang Y and Le H: Expression of miR-29c, miR-93, and miR-429
as potential biomarkers for detection of early stage non-small lung
cancer. PLoS One. 9:e877802014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miyazono K: Transforming growth
factor-beta signaling in epithelial-mesenchymal transition and
progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci.
85:314–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu D, Zhou Y, Pan H, Zhou J, Fan Y and Qu
P: microRNA-99a inhibiting cell proliferation, migration and
invasion by targeting fibroblast growth factor receptor 3 in
bladder cancer. Oncol Lett. 7:1219–1224. 2014.PubMed/NCBI
|
|
25
|
Hu X, Macdonald DM, Huettner PC, Feng Z,
El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN and Wang X:
A miR-200 microRNA cluster as prognostic marker in advanced ovarian
cancer. Gynecol Oncol. 114:457–464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xie P, Xu F, Cheng W, Gao J, Zhang Z, Ge
J, Wei Z, Xu X and Liu Y: Infiltration related miRNAs in bladder
urothelial carcinoma. J Huazhong Univ Sci Technolog Med Sci.
32:576–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nam EJI, Yoon H, Kim SW, Kim H, Kim YT,
Kim JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun T, Wang C, Xing J and Wu D: miR-429
modulates the expression of c-myc in human gastric carcinoma cells.
Eur J Cancer. 47:2552–2559. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun Y, Shen S, Tang H, Xiang J, Peng Y,
Tang A, Li N, Zhou W, Wang Z, Zhang D, et al: miR-429 identified by
dynamic transcriptome analysis is a new candidate biomarker for
colorectal cancer prognosis. OMICS. 18:54–64. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu X, Liu Y, Wu S, Shi X, Li L, Zhao J
and Xu H: Tumor-suppressing effects of miR-429 on human
osteosarcoma. Cell Biochem Biophys. 70:215–224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye ZB, Ma G, Zhao YH, Xiao Y, Zhan Y, Jing
C, Gao K, Liu ZH and Yu SJ: miR-429 inhibits migration and invasion
of breast cancer cells in vitro. Int J Oncol. 46:531–538.
2015.PubMed/NCBI
|
|
32
|
Wang Y, Li M, Zang W, Ma Y, Wang N, Li P,
Wang T and Zhao G: MiR-429 up-regulation induces apoptosis and
suppresses invasion by targeting Bcl-2 and SP-1 in esophageal
carcinoma. Cell Oncol (Dordr). 36:385–394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dynan WS and Tjian R: The
promoter-specific transcription factor Sp1 binds to upstream
sequences in the SV40 early promoter. Cell. 35:79–87. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang WC and Hung JJ: Functional role of
post-translational modifications of Sp1 in tumorigenesis. J Biomed
Sci. 19:942012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kadonaga JT, Courey AJ, Ladika J and Tjian
R: Distinct regions of Sp1 modulate DNA binding and transcriptional
activation. Science. 242:1566–1570. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Philipsen S and Suske G: A tale of three
fingers: The family of mammalian Sp/XKLF transcription factors.
Nucleic Acids Res. 27:2991–3000. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Narayan VA, Kriwacki RW and Caradonna JP:
Structures of zinc finger domains from transcription factor Sp1.
Insights into sequence-specific protein-DNA recognition. J Biol
Chem. 272:7801–7809. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and krüppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Beishline K and Azizkhan-Clifford J: Sp1
and the ‘Hallmarks of Cancer’. FEBS J. 282:224–258. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sankpal UT, Goodison S, Abdelrahim M and
Basha R: Targeting Sp1 transcription factors in prostate cancer
therapy. Med Chem. 7:518–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yue L, Li L, Liu F, Hu N, Zhang W, Bai X,
Li Y, Zhang Y, Fu L, Zhang X and Ye L: The oncoprotein HBXIP
activates transcriptional coregulatory protein LMO4 via Sp1 to
promote proliferation of breast cancer cells. Carcinogenesis.
34:927–935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bonofiglio D, Qi H, Gabriele S, Catalano
S, Aquila S, Belmonte M and Andò S: Peroxisome
proliferator-activated receptor gamma inhibits follicular and
anaplastic thyroid carcinoma cells growth by upregulating
p21Cip1/WAF1 gene in a Sp1-dependent manner. Endocr Relat Cancer.
15:545–557. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yin P, Zhao C, Li Z, Mei C, Yao W, Liu Y,
Li N, Qi J, Wang L, Shi Y, et al: Sp1 is involved in regulation of
cystathionine gamma-lyase gene expression and biological function
by PI3K/Akt pathway in human hepatocellular carcinoma cell lines.
Cell Signal. 24:1229–1240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang C and Xie K: Crosstalk of Sp1 and
Stat3 signaling in pancreatic cancer pathogenesis. Cytokine Growth
Factor Rev. 23:25–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pathi S, Jutooru I, Chadalapaka G, Nair V,
Lee SO and Safe S: Aspirin inhibits colon cancer cell and tumor
growth and downregulates specificity protein (Sp) transcription
factors. PLoS One. 7:e482082012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang
X, Jiang L, Sun Z, Miao Z and Xu H: MicroRNA-335 acts as a
metastasis suppressor in gastric cancer by targeting Bcl-w and
specificity protein 1. Oncogene. 31:1398–1407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang YT, Chuang JY, Shen MR, Yang WB,
Chang WC and Hung JJ: Sumoylation of specificity protein 1 augments
its degradation by changing the localization and increasing the
specificity protein 1 proteolytic process. J Mol Biol. 380:869–885.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC,
Chang WC and Hung JJ: Sp1 expression regulates lung tumor
progression. Oncogene. 31:3973–3988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Trisciuoglio D, Iervolino A, Candiloro A,
Fibbi G, Fanciulli M, ZangemeisterWittke U, Zupi G and Del Bufalo
D: Bcl-2 induction of urokinase plasminogen activator receptor
expression in human cancer cells through Sp1 activation:
involvement of ERK1/ERK2 activity. J Biol Chem. 279:6737–6745.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
DeSiervi A, Marinissen M, Diggs J, Wang
XF, Pages G and Senderowicz A: Transcriptional activation of p21
(waf1/cip1) by alkylphospholipids: Role of the mitogen-activated
protein kinase pathway in the transactivation of the human
p21(waf1/cip1) promoter by Sp1. Cancer Res. 64:743–750. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang H, Li M, Zhang R, Wang Y, Zang W, Ma
Y, Zhao G and Zhang G: Effect of miR-335 upregulation on the
apoptosis and invasion of lung cancer cell A549 and H1299. Tumour
Biol. 34:3101–3109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jiang J, Lv X, Fan L, Huang G, Zhan Y,
Wang M and Lu H: MicroRNA-27b suppresses growth and invasion of
NSCLC cells by targeting Sp1. Tumour Biol. 35:10019–10023. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mao Y, Chen H, Lin Y, Xu X, Hu Z, Zhu Y,
Wu J, Xu X, Zheng X and Xie L: microRNA-330 inhibits cell motility
by downregulating Sp1 in prostate cancer cells. Oncol Rep.
30:327–333. 2013.PubMed/NCBI
|
|
54
|
Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F,
Shen Y, Lu W, Wan X and Xie X: miR-375 is down-regulated in
squamous cervical cancer and inhibits cell migration and invasion
via targeting transcription factor SP1. Am J Pathol. 179:2580–2588.
2011. View Article : Google Scholar : PubMed/NCBI
|