Introduction
Sirtuins, homologues of the Saccharomyces
cerevisiae silent information regulator 2 (Sir2) protein, are
nicotinamide adenine dinucleotide (NAD+)-dependent,
class III histone deacetylases (HDACs), conserved from bacteria to
humans (1). In total, 7 types of
sirtuins (Sirt-1-7) have been identified in mammals, with each of
them containing a conserved 275-amino acid catalytic domain and a
NAD+ binding domain (2).
Sirtuins may be located in different subcellular compartments,
including the nucleus (Sirt-6 and Sirt-7) and mitochondrion
(Sirt-3, Sirt-4 and Sirt-5), while certain sirtuins (Sirt-1 and
Sirt-2) shuttle between the nucleus and cytoplasm (3–5). Certain
sirtuins may re-localize depending on the cell or tissue type,
developmental stage, stress condition and metabolic status,
suggesting that their localization is important for the regulation
of their activity (6). Sirtuins are
involved in various cellular functions, such as chromosomal
stability and gene expression, cell proliferation and survival,
cell cycle progression, cell division control, cell death, cell
metabolism and calorie restriction (1). In detail, sirtuins may affect gene
expression epigenetically by histone acetylation (7). Through acetylation, histones accumulate
negative charges that decrease the interaction of their N-terminal
regions with the negatively charged phosphate groups of DNA. As a
consequence, chromatin is transformed into a more relaxed structure
that permits gene transcription. De-acetylation, conversely, causes
chromatin condensation that reduces the level of transcriptional
activity.
Over the past decade, sirtuins have emerged as the
principal factor in sensing and regulating cellular response to
oxidative and metabolic stress (1,6,8,9). These
proteins are involved in cell senescence, cell differentiation,
stress-induced apoptosis cell growth, angiogenesis and blood supply
(8). In addition, the epigenetic
regulation of sirtuins has been characterized in numerous types of
cancers, including lymphomas, leukemia, prostate, breast, intestine
and colon cancers, and the effect of sirtuins on the expression of
proteins involved in tumorigenesis, such as p53, p73, nuclear
factor-κB (NF-κB) and forkhead box subgroup O (FOXO), has been well
studied (9–15).
The ‘double-face’ of Sirt-1 in tumorigenesis was
recently reported (16). It appears
that Sirt-1 can inhibit inflammation and multistage carcinogenesis
by acting as a tumor suppressor (15,16).
Paradoxically, it may also accelerate tumorigenesis via multiple
mechanisms, such as inactivation of tumor suppressors, activation
of oncoproteins and development of a microenvironment favorable to
tumor survival (16). Accordingly,
studies have shown that Sirt-1 may be either upregulated (17,18) or
downregulated in human gliomas (19,20). Qu
et al (21) demonstrated that
Sirt-1 knockdown significantly delays mitotic entry of glioma
cells, inhibits their growth and proliferation and promotes
apoptosis via the phosphatase and tensin homolog
(PTEN)/phosphoinositide 3-kinase (PI3K)/AKT signaling pathway,
suggesting that Sirt-1 may be a promoter of tumorigenesis in glioma
through the PTEN/PI3K/AKT signaling pathway (21). By contrast, it was recently shown that
Sirt-1 expression levels are lower in glioblastoma, bladder,
prostate and ovarian carcinomas compared to normal controls
(20,22). In addition, previous studies strongly
suggest critical roles for microRNAs (miRNAs) in regulating Sirt-1
(23–25). miRNAs are short non-coding endogenous
RNAs that post-transcriptionally regulate the expression of a large
number of target genes involved in physiological and pathological
pathways. miRNAs are considered to play important roles in cancer
by regulating the expression of various oncogenes and tumor
suppressors (26), and alteration of
miRNA profiles has been demonstrated in glioma tissues (26–28).
Although the pattern of Sirt-1 expression and its
relevance has been reported in several types of cancer, to the best
of our knowledge, there are no studies investigating the expression
pattern of Sirt-1 in human glioma tissues and primary glioma cell
lines, and the role of Sirt-1 in gliomagenesis remains largely
unknown (11–15). In the present study, to better
characterize the role of Sirt-1 in human gliomas, the miRNA
expression of Sirt-1 and its associated regulators was investigated
in astrocytic tumors with different grades of malignancy [World
Health Organization (WHO) II–IV] (29), and also in primary human glioma cell
lines for the first time.
Materials and methods
Tissue samples
The Institutional Review Board of the Department of
Neurosciences of the University of Messina (Messina, Italy)
discussed and approved the current study. Written informed consent
was obtained from all subjects or their caregivers at the moment of
diagnostic procedures, including for research purposes, according
to the Declaration of Helsinki. In total, 30 tumor specimens of
astrocytomas of different histological grades were obtained from
adult patients who had undergone craniotomy for microsurgical brain
tumor removal at the Neurosurgery Unit of the University of Messina
between 2011 and 2014. Samples were immediately frozen in liquid
nitrogen and stored at −80°C for biomolecular analyses and formalin
fixed for subsequent histological evaluation.
No patients received neoadjuvant therapy
(radiotherapy and/or chemotherapy). According to the revised WHO
classification (29), tumors were
histologically classified as: Diffuse fibrillary astrocytoma [low
grade astrocytoma (LGA)] (n=10); anaplastic astrocytomas (AA)
(n=10); and glioblastoma multiforme (GBM) (n=10). Furthermore,
samples of non-neoplastic brain tissue (NBT) (n=5) derived from the
temporal lobes of patients who underwent brain surgery at the
Neurosurgery Unit of the University of Messina between 2011 and
2014 for cerebral hemorrhage, including histologically verified
normal cortex and white matter, were used as controls.
Cell lines
Primary glioma cell lines and normal human
astrocytes (NHAs) were used as a control for the present study. The
human glioma cell lines were obtained from samples of grade II, III
and IV glioma, as follows: PLGAC, primary LGA cells (grade II);
PAAC, primary AA cells (grade III); and PGBMC, primary GBM cells
(grade IV). Briefly, fresh tumor samples were washed extensively
with phosphate-buffered saline and were processed by mincing the
tissue with operative scissors, followed by enzymatic digestion
with 150 U/ml hyaluronidase and 250 U/ml collagenase type IV
(Sigma-Aldrich, St. Louis, MO, USA). The single-cell suspension was
filtered through a 100-µm cell strainer, washed and then plated in
at a density of 5×104 cells in DMEM medium supplemented
with 10% FCS, penicillin at 100 U/ml, streptomycin at 100 µg/ml and
2 mM L-glutamine (Lonza, Verviers, Belgium) at 37°C in 5%
CO2. The presence of glioma cells was assessed by
staining proliferating cells with a monoclonal mouse anti-glial
fibrillary acidic protein antibody (#M0761; Dako, Glostrup,
Denmark; dilution, 1:100) and analyzing ploidy by
fluorescence-activated cell sorting analysis.
Western blotting
Tumor specimens were homogenized on ice in lysis
buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid;
1 mM ethylenediaminetetraacetic acid (EDTA); 2 mM ethylene glycol
tetraacetic acid (EGTA); 150 mm NaCl; 0.1% sodium dodecyl sulfate
(SDS); 1% Nonidet P-40; 0.5% deoxycholate acid, pH 7.5; a mixture
of protease inhibitors; 0.5 mm phenylmethylsulfonyl fluoride; 10
µg/ml aprotinin; 10 µg/ml leupeptin; and 10 µg/ml pepstatin),
purchased from Sigma-Aldrich. Cell lines were lysed in
radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH 7.4; 150
mM NaCl; 1 mM EDTA; 1 mM EGTA; 2% Triton), including proteinase
inhibitors. The concentrations of protein were determined by the
Lowry method. Protein samples (30 µg) were loaded onto 8%
SDS-polyacrylamide gel electrophoresis and transferred onto
methanol-activated polyvinylidene difluoride membrane. Membranes
were incubated for 1 h in a solution containing 1% bovine serum
albumin (Bio-Rad Laboratories, Inc., Milan, Italy) in Tris-buffered
saline with Tween 20 (Sigma-Aldrich), then incubated overnight with
mouse monoclonal antibodies against Sirt-1 (clone 1F3; #sc-74465;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA; dilution, 1:500)
and β-actin (#sc-8432; Santa Cruz Biotechnology, Inc.; dilution,
1:1,000). The membranes were incubated with horseradish
peroxidase-conjugated rabbit anti-mouse immunoglobulin G antibody
(#P0260; Dako Italia Srl, Milan, Italy; dilution, 1:5,000) for 2 h.
Immunoreactive bands were visualized using the ECL
Chemiluminescence kit (Amersham; GE Healthcare Life Sciences,
Chalfont, UK).
Semi-quantitative evaluation
Semi-quantitative evaluation of protein levels
detected by immunoblotting was performed with computer-assisted
densitometry (UN-SCAN-IT gel version 6.1; Silk Scientific, Inc.,
Orem, UT, USA). Different exposure times were used for each blot.
Protein amounts were normalized to the β-actin signal. Data were
acquired and integrated density values were expressed as a
percentage of densitometric levels using arbitrary densitometric
units.
RNA, miRNA isolation and cDNA
synthesis
Total RNA was extracted from specimens of glioma
tissues and cell cultures using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA USA), according to the
manufacturer's instructions. Total RNA concentration and integrity
were checked using an Agilent Bioanalyzer (Agilent Technologies,
Inc., Santa Clara, CA, USA). Subsequently, 300 ng of total mRNA per
sample was reverse transcribed (RT) into cDNA using the High
Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.).
miRNAs were extracted from glioma tissue and cell
lines using the miRVana Isolation kit (Ambion; Thermo Fisher
Scientific, Inc.), following the manufacturer's protocol. The
enriched miRNAs fraction was converted in cDNA using the TaqMan
MicroRNA Reverse Transcriptase kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.).
In total, 2 µl of cDNA was used for specific miRNA
TaqMan assay, which was amplified using Applied Biosystems TaqMan
MicroRNA assay (hsa-miR-34a, hsa-miR-217 and hsa-miR-132; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. All RT reactions, including no-template controls and
RT controls, were run in triplicate.
RT-quantitative polymerase chain
reaction (qPCR) of Sirt-1 and miRNAs
RT-qPCR for Sirt-1 was performed using a standard
TaqMan PCR kit procedure on an AB-7300 RT-PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.).
For miRNA quantification, 2 µl of cDNA was used for
each specific miRNA TaqMan assay (hsa-miR-34a, hsa-miR-217 and
hsa-miR-132) according to the manufacturer's instructions. All RT
reactions, including no-template controls and RT controls, were
performed in triplicate. RNU6 small nuclear RNA was used to
normalize miRNA expression levels owing to its claimed expression
stability and its wide use as loading control in several published
miRNA expression studies (28,30,31).
Relative fold expression and changes were calculated using
2−ΔΔCt method. The expression levels of miRNAs are
indicated as either fold expression (<0.3 downregulation and
>3 upregulation) compared to NBT or NHA. Results were
represented as Log10RQ.
Target prediction tools
miRNAs that target Sirt-1 were identified by
examining the Sirt-1 3′-untranslated region (UTR) with
bioinformatics algorithms that predict miRNA target sites (32). Specifically, four online databases,
miRDB (http://mirdb.org/miRDB/), TargetScan
(www.targetscan.org), microRNA.org (www.microrna.org) and PicTar (http://pictar.mdc-berlin.de), were used for the
analysis of the alignment between miRNAs and the 3′-UTR of
Sirt-1.
Statistical data analysis
Statistical analysis was performed using INSTAT,
version 3.0, and PRISM, version 4.0 (GraphPad, San Diego, CA, USA).
Unpaired t-test with Welch correction was used to compare Sirt-1
expression levels. P<0.05 was considered to indicate a
statistically significant difference.
Results
Downregulation of Sirt-1 in glioma
tissues and cell lines
Sirt-1 protein expression was downregulated in
high-grade tumors (AAs and GBMs) compared to LGAs and controls
(NBTs), as shown in Fig. 1 (LGA and
AA vs. NBT, P<0.01; GBM vs. NBT, P<0.001). In addition, a
similar pattern of Sirt-1 expression was observed in glioma cell
lines compared to NHAs (P<0.0001; Fig.
1B).
 | Figure 1.(A) Western blot analysis of Sirt-1
in NBT, LGA, AA and GBM tissues. Representative autoradiography
shows Sirt-1 and β-actin expression. Quantitative data indicate the
mean ± standard deviation (error bars). *P<0.01, LGA and AA vs.
NBT; **P<0.001, GBM vs. NBT. (B) Western blot analysis of Sirt-1
on primary glioma cell cultures. Representative autoradiography
shows Sirt-1 expression and β-actin in NHA, primary LGA cells
(grade II), primary AA cells (grade III) and primary GBM cells
(grade IV). Quantitative data indicate the mean ± standard
deviation (error bars). *P<0.001 vs. NHA. Sirt-1, sirtuin-1;
NBT, neoplastic brain tissue; LGA, low grade astrocytoma; AA,
anaplastic astrocytoma; GBM, glioblastoma multiforme; NHA, normal
human astrocyte. |
To better understand the involvement of Sirt-1 in
glioma development and progression, Sirt-1 mRNA levels were also
analyzed by RT-qPCR in human NBTs, LGAs (WHO grade II), AAs (grade
III) and GBMs (grade IV) and in primary glioma cell lines: PLGAC
(primary LGA cells); PAAC (primary AA cells); and PGBMC (primary
GBM cells). RT-qPCR analysis of glioma samples was consistent with
the Sirt-1 protein levels revealed by western blotting. In
particular, Sirt-1 mRNA levels in high-grade glioma (AA and GBM)
were significantly lower than in low-grade tissues (LGA) as
compared to NBT (LGA vs. NBT, P>0.05; AA vs. NBT, P<0.0001;
GBM vs. NBT, P<0.01; Fig. 2A).
In primary LGA and AA cells, reduced expression of
Sirt-1 was observed, according to the results obtained for protein
expression in the same cells. Notably, in primary GBM cells a
different expression pattern was observed, with an upregulation of
Sirt-1 (GBM cells vs. NHA, P<0.0001; Fig. 2B).
Expression of miR-132, miR-34a and
miR-217
Furthermore, the level of expression of miR-132,
miR-34a and miR-217 was quantified, which were chosen by analyzing
the alignment details of the 3′-UTR region of Sirt-1 with a
computational miRNA target prediction tool (www.microRNA.org/) and comparing the results with
validated pathways (Fig. 3). miR-217
(P<0.001 vs. LGA and II primary cells; Fig. 3A and B) and miR-34a (P<0.001 vs.
LGA and PGLAC (grade II) cells; Fig. 3E
and F) were consistently upregulated in high-grade glioma and
tumorous cell lines, while a downregulation of miR-132 (P<0.001
vs. LGA and PGLAC (grade II) cells; Fig.
3C and D) was observed in high-grade glioma cell lines. These
data suggest that the assessed miRNAs inversely modulate Sirt-1
expression, depending on the glioma grade. Analysis of Sirt-1 and
miRNA expression may be a valuable marker of glioma
progression.
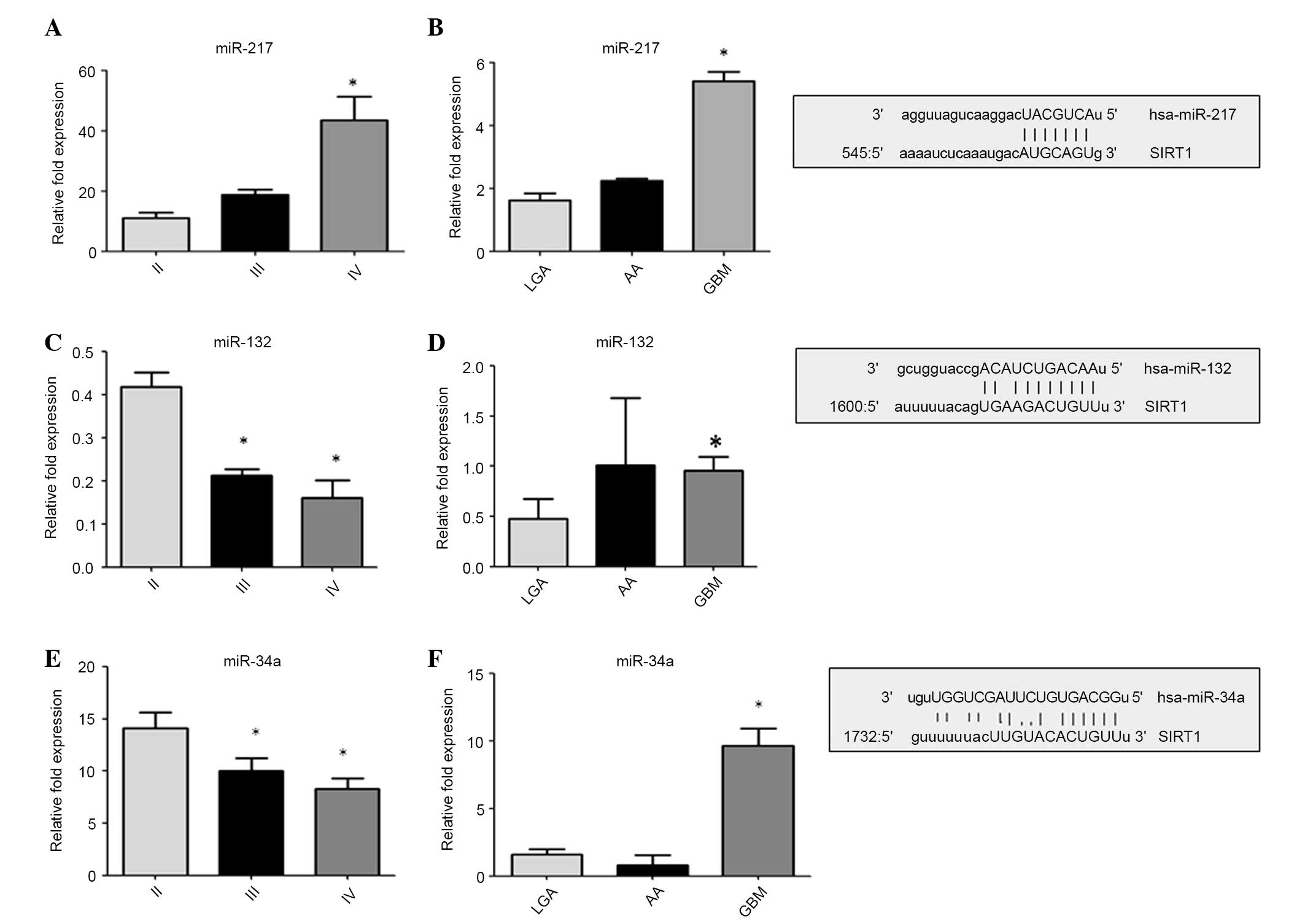 | Figure 3.Expression of miR-132, miR-34a and
miR-217. miRNAs expression were quantified by reverse
transcription-quantitative polymerase chain reacion in primary
glioma cell lines (panels A, C and E) and in tumor tissues (panels
B, D and F). The data are expressed as the log10 RQ relative to U6.
On the right: Aligment details with 3′-UTR region of SIRT-1. Sirt-1
is direct target of miR-34a, miR-217 and miR-132. The predicted
highly conserved miRNAs targeting sequence located at the 3′ UTR of
Sirt-1 mRNA found on microRNA.org.
Sirt-1, sirtuin-1; LGA, low grade astrocytoma; AA, anaplastic
astrocytoma; GBM, glioblastoma multiforme; miR, microRNA; 3′-UTR,
3′-untranslated region. |
Discussion
Previous studies have demonstrated that Sirt-1 is
overexpressed in tumors such as lymphomas (11), leukemia (12), and prostate (13), breast (14) and colon cancers (15).
Usually, Sirt-1 is located in the nucleus, bound to
histones H1, H3 and H4 (8,33). Similar to yeast Sir2, human Sirt-1
remodels chromatin through histone deacetylation and silences the
transcription of numerous genes by heterochromatin formation
(3–5,8,34). The genes repressed by this mechanism
at their promoters include: E-cadherin; mismatch repair gene mutL
homolog 1; transcription factors GATA-4 and GATA-5; p27; and
cellular retinoid binding protein 1 (35). All these genes are actively involved
in tumorigenesis, progression and metastasis (35,36).
Furthermore, Sirt-1 deacetylases non-histone
substrates in cancer cells, such as the following transcription
factors: p53; B-cell lymphoma-associated X protein, NF-κB, E2F1,
FOXO1 (37,38), components of the core RNA polymerase I
and the histone acetyltransferase p300/cAMP response element
binding protein binding protein (39), thereby reducing gene expression
(40). Other non-histone substrates
include signaling factors, such as endothelial nitric oxide
synthase, and DNA repair proteins, such as Ku-70 (36). Although the role of Sirt-1 it has been
reported in a variety of cancers, to the best of our knowledge,
there are no studies investigating its expression pattern in human
glioma tissues and primary glioma cell lines of different
grades.
The present results showed that Sirt-1 expression is
reduced in human astrocytic tumors, with a more prominent
downregulation in high-grade tumors (AAs and GBMs) compared with
non-neoplastic brain tissue, thus suggesting that Sirt-1 expression
is correlated with glioma tumor stages. In addition, the expression
level of Sirt-1 mRNA in primary glioma cell lines is upregulated in
high-grade PGBMCs as compared with normal human astrocytes (NHA).
This different expression between high-grade tissues and and
high-grade primary GBM cells suggests that Sirt-1 mRNA may undergo
regulation at the transcriptional and translational levels, with
more discordant expression patterns and protein levels, or the
difference may be due to epigenetic effects on Sirt-1 mRNA or the
variability of the microenvironment of brain cancer, leading to
destabilization of the mRNA product or translational repression
(41). The apparently controversial
role of Sirt-1 may depend on the presence of other molecules
regulating Sirt-1 during glioma progression or on the different
cell-context specificity (immortalized cell cultures vs. human
glioma cells). Among these molecules, microRNAs could play a key
role. In fact, certain miRNAs are overexpressed in tumors and act
as oncogenes, promoting carcinogenesis by targeting tumor
suppressors. Other miRNAs are downregulated in tumors and generally
participate in oncogene overexpression. Additionally, under
baseline conditions, miRNAs appear to act as moderate regulators of
gene expression, but under conditions of stress or disease, miRNAs
appear to exert more pronounced functions (23).
Overall, >16 miRNAs regulate Sirt-1 expression
and activity. Among these miRNAs, miR-34a has been the most
studied. miR-34a was identified as a tumor suppressor gene in
neuroblastoma (42). One mechanism by
which miR-34a expression decreases in cancers is aberrant CpG
methylation of the promoter of miR-34a (43). Reduced expression of miR-34a alters
cellular function in cancer cells. Ectopic expression of miR-34a
induces cell cycle arrest in the G1 phase and apoptosis in several
cancer cells. Numerous potential targets of miR-34a have been
identified (44–46). However, the precise molecular targets
of miR-34a remain unknown. Sirt-1 regulation by miR-34a has been
reported in colorectal cancer, prostate cancer, hepatocarcinoma,
glioma and pancreatic cancer (48–52).
miRNA-132 affects Sirt-1 regulation in adipocytes.
Overexpression of miR-132 decreases Sirt-1 expression, which blocks
deacetylation of p65 NF-κB (23).
Proliferation and colony formation of pancreatic cancer cells was
suppressed in cells transfected with miR-132 mimics and enhanced in
cells transfected with miR-132 inhibitor by negatively regulating
the Akt-signaling pathway (53).
Computational target prediction identified homology
between miR-217 and 3′-UTR of human Sirt-1 mRNA. In a previous
study comparing the expression of miR-217 during the aging process,
from young to senescent endothelial cells, overexpression of
miR-217 suppressed Sirt-1 expression in young endothelial cells as
compared with senescent cells (54).
By contrast, inhibition of miR-217 increases Sirt-1 expression and
promotes cellular senescence in older human umbilical vein
endothelial cells. This study confirms that miR-217 level is
negatively associated with Sirt-1 expression (54).
One of the most notable aspects of miRNA biology is
that one miRNA often regulates multiple genes and one gene is
modulated by various miRNAs (55).
In the present study, data obtained suggest that
miR-34a and miR-217 upregulation in high-grade tissue and cells is
responsible for Sirt-1 downregulation, while miR-132 downregulation
may be the cause of Sirt-1 overexpression in high grade primary
cell lines. This intriguing profile of expression between gliomas
tissues and cells may be due to a number of factors. As
aforementioned, regulation of Sirt-1 mRNA expression may be
affected at the transcriptional and translational level or may be
associated with various microenvironmental conditions. In addition,
it is well known that miRNAs regulate gene expression by
interacting with multiple networks and complex mechanisms, and this
may account for the different profile of expression between human
glioma tissues and cells. Indeed, every miRNA target contains miRNA
response elements (MREs), to which miRNAs are attracted. At this
level, the activity of the miRNAs may be modulated by the presence
of competitive endogenous RNAs that, acting as sponges of miRNAs,
vie for the miRNAs with shared MREs and affect the available levels
of miRNAs in a cell, thus modulating the expression of target
proteins (56–59). However, miRNAs may also activate gene
expression, switching between repression and stimulation in
response to different cell types, specific cellular and
environmental conditions, and cofactors, which allows the cells to
rapidly adapt to a variety of stimuli (60–62). The
ambiguous effects of miRNAs make it challenging to study and fully
understand the mechanisms by which miRNAs regulate gene
expression.
The role of Sirt-1 in cancer is currently under
debate due to numerous controversial findings suggesting that
Sirt-1 acts as an oncosuppressor or as an oncogene (16,22,63). Few
data specifically addressing the role of Sirt-1 in astrocytic
tumors are currently available. Whereas certain studies reveal that
overexpression of Sirt-1 is associated with gliomagenesis and its
downregulation induces apoptosis and increases radiosensitivity in
cancer cells, other studies show low levels of expression of Sirt-1
in gliomas (17,18,20). In
particular, Liu et al (18)
found that in GBM stem cells, Sirt-1 expression is 4.9-fold higher
in poorly differentiated, CD133-positive tumor cells than in more
differentiated CD133-negative tumor cells. Chang et al
(17) reported that the knockdown of
Sirt-1 expression in GBM stem cells enhances radiosensitivity and
apoptotic activity in vitro and in vivo, suggesting
that Sirt-1 could be a key regulator of GBM resistance to
radiotherapy.
Wang et al (20) revealed a decreased expression level in
GBM, although this result was not obtained by direct measurements,
but rather by indirect analysis of public microarray datasets. The
same decrement of Sirt-1 expression was observed by Lages et
al (19) in GBM human samples and
was associated with the different deregulation of Sirt-1 as an
miRNA target. These data agree with the present findings of Sirt-1
downregulation in human astrocytic tumors.
To the best of our knowledge, the present study is
the only study that has analyzed the expression of Sirt-1 in human
samples of glioma with different grades of malignancy and Sirt-1
level expression in primary human glioma cell lines.
Whether Sirt-1 acts as an oncogene or tumor
suppressor remains to be determined. However, the ability of Sirt-1
to protect the cell and expand the life span of the cell is
evident. The unclear role of Sirt-1 may depend on the presence of
other molecules, such as miRNAs, regulating Sirt-1 during glioma
progression or on the different cell-context specificity
(immortalized cell cultures vs. human glioma cells).
In conclusion, the present results support the
important role for Sirt-1 as intrinsic regulator of tumor
progression. However, the biological functions of Sirt-1 and its
complex molecular pathways in gliomagenesis require additional
investigation, with analysis of Sirt-1 gene silencing and
functional analysis of involved miRNAs, for an in-depth
characterization of the role of Sirt-1 in glioma development and
progression.
Acknowledgements
The present study was supported by National
Operative Programme (grant no., 02_00643_3604826; Ministry of
Education, Universities and Research, Rome, Italy).
References
|
1
|
Harting K and Knöll B: SIRT2-mediated
protein deacetylation: An emerging key regulator in brain
physiology and pathology. Eur J Cell Biol. 89:262–269. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sherman JM, Stone EM, FreemanCook LL,
Brachmann CB, Boeke JD and Pillus L: The conserved core of a human
SIR2 homologue functions in yeast silencing. Mol Biol Cell.
10:3045–3059. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blander G and Guarente L: The Sir2 family
of protein deacetylases. Annu Rev Biochem. 73:417–435. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taylor DM, Maxwell MM, LuthiCarter R and
Kazantsev AG: Biological and potential therapeutic roles of sirtuin
deacetylases. Cell Mol Life Sci. 65:4000–4018. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McGuinness D, McGuinness DH, McCaul JA and
Shiels PG: Sirtuins, bioageing, and cancer. J Aging Res.
2011:2357542011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haberland M, Montgomery RL and Olson EN:
The many roles of histone deacetylases in development and
physiology: Implications for disease and therapy. Nat Rev Genet.
10:32–42. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Finkel T, Deng CX and Mostoslavsky R:
Recent progress in the biology and physiology of sirtuins. Nature.
460:587–591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Haigis MC and Sinclair DA: Mammalian
sirtuins: Biological insights and disease relevance. Annu Rev
Pathol. 5:253–295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Miremadi A, Oestergaard MZ, Pharoah PD and
Caldas C: Cancer genetics of epigenetic genes. Hum Mol Genet.
16:R28–R49. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jang KY, Hwang SH, Kwon KS, Kim KR, Choi
HN, Lee NR, Kwak JY, Park BH, Park HS, Chung MJ, et al: SIRT1
expression is associated with poor prognosis of diffuse large
B-cell lymphoma. Am J Surg Pathol. 32:1523–1531. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bradbury CA, Khanim FL, Hayden R, Bunce
CM, White DA, Drayson MT, Craddock C and Turner BM: Histone
deacetylases in acute myeloid leukemia show a distinctive pattern
of expression that changes selectively in response to deacetylase
inhibitors. Leukemia. 19:1751–1759. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huffman DM, Grizzle WE, Bamman MM, Kim JS,
Eltoum IA, Elgavish A and Nagy TR: SIRT1 is significantly elevated
in mouse and human prostate cancer. Cancer Res. 67:6612–6618. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McGlynn L, Curle J, Edwards J and Shies P:
Evaluating the role of sirtuins 5,6 & 7 in breast cancer.
Cancer Res. 69:30342009. View Article : Google Scholar
|
|
15
|
Firestein R, Blander G, Michan S,
Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S,
de Cabo R, Fuchs C, et al: The SIRT1 deacetylase suppresses
intestinal tumorigenesis and colon cancer growth. PLoS One.
3:e20202008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song NY and Surh YJ: Janus-faced role of
SIRT1 in tumorigenesis. Ann N Y Acad Sci. 1271:10–19. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang CJ, Hsu CC, Yung MC, Chen KY, Tzao
C, Wu WF, Chou HY, Lee YY, Lu KH, Chiou SH, et al: Enhanced
radiosensitivity and radiation-induced apoptosis in glioma
CD133-positive cells by knockdown of SirT1 expression. Biochem
Biophys Res Commun. 380:236–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu G, Yuan X, Zeng Z, Tunici P, Ng H,
Abdulkadir IR, Lu L, Irvin D, Black KL and Yu JS: Analysis of gene
expression and chemoresistance of CD133+ cancer stem cells in
glioblastoma. Mol Cancer. 5:672006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lages E, Guttin A, ElAtifi M, Ramus C,
Ipas H, Dupré I, Rolland D, Salon C, Godfraind C, de Fraipont F, et
al: MicroRNA and target protein patterns reveal physiopathological
features of glioma subtypes. PLoS One. 6:e206002011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang RH, Sengupta K, Li C, Kim HS, Cao L,
Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al: Impaired DNA damage
response, genome instability, and tumorigenesis in SIRT1 mutant
mice. Cancer Cell. 14:312–323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu Y, Zhang J, Wu S, Li B, Liu S and Cheng
J: SIRT1 promotes proliferation and inhibits apoptosis of human
malignant glioma cell lines. Neurosci Lett. 525:168–172. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng CX: SIRT1, Is it a tumor promoter or
tumor suppressor? Int J Biol Sci. 5:147–152. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yamakuchi M: MicroRNA regulation of SIRT1.
Front Physiol. 3:682012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Wang WY, Mao YW, Graff J, Guan JS,
Pan L, Mak G, Kim D, Su SC and Tsai LH: A novel pathway regulates
memory and plasticity via SIRT1 and miR-134. Nature. 466:1105–1109.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamakuchi M and Lowenstein C J: MiR-34,
SIRT1 and p53: the feedback loop. Cell Cycle. 8:712–715. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ciafrè SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jia Z, Wang K, Zhang A, Wang G, Kang C,
Han L and Pu P: miR-19a and miR-19b overexpression in gliomas.
Pathol Oncol Res. 19:847–853. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Conti A, Aguennouz M, La Torre D,
Tomasello C, Cardali S, Angileri FF, Maio F, Cama A, Germanò A,
Vita G and Tomasello F: miR-21 and 221 upregulation and miR-181b
downregulation in human grade II–IV astrocytic tumors. J
Neurooncol. 93:325–332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–52. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song J, Bai Z, Han W, Zhang J, Meng H, Bi
J, Ma X, Han S and Zhang Z: Identification of suitable reference
genes for qPCR analysis of serum microRNA in gastric cancer
patients. Dig Dis Sci. 57:897–904. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nakahata Y, Sahar S, Astarita G, Kaluzova
M and Sassone-Corsi P: Circadian control of the NAD+ salvage
pathway by CLOCK-SIRT1. Science. 324:654–657. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vaquero A, Scher M, Lee D,
ErdjumentBromage H, Tempst P and Reinberg D: Human SirT1 interacts
with histone H1 and promotes formation of facultative
heterochromatin. Mol Cell. 16:93–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pruitt K, Zinn RL, Ohm JE, McGarvey KM,
Kang SH, Watkins DN, Herman JG and Baylin SB: Inhibition of SIRT1
reactivates silenced cancer genes without loss of promoter DNA
hypermethylation. PLoS Genet. 2:e402006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu T, Liu PY and Marshall GM: The
critical role of the class III histone deacetylase SIRT1 in cancer.
Cancer Res. 69:1702–1705. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saunders LR and Verdin E: Sirtuins:
Critical regulators at the crossroads between cancer and aging.
Oncogene. 26:5489–5504. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2 (SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bouras T, Fu M, Sauve AA, Wang F, Quong
AA, Perkins ND, Hay RT, Gu W and Pestell RG: SIRT1 deacetylation
and repression of p300 involves lysine residues 1020/1024 within
the cell cycle regulatory domain1. J Biol Chem. 280:10264–10276.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Muth V, Nadaud S, Grummt I and Voit R:
Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA
polymerase I transcription. EMBO J. 20:1353–1362. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Placone AL, Quiñones-Hinojosa A and
Searson PC: The role of astrocytes in the progression of brain
cancer: Complicating the picture of the tumor microenvironment.
Tumour Biol. 2015.(Epub ahead of print). PubMed/NCBI
|
|
42
|
Dutta KK, Zhong Y, Liu YT, Yamada T,
Akatsuka S, Hu Q, Yoshihara M, Ohara H, Takehashi M, Shinohara T,
et al: Association of microRNA-34a overexpression with
proliferation is cell type-dependent. Cancer Sci. 98:1845–1852.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lodygin D, Tarasov V, Epanchintsev A,
Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J and Hermeking
H: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bommer GT, Gerin I, Feng Y, Kaczorowski
AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al:
p53-mediated activation of miRNA34 candidate tumor-suppressor
genes. Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tarasov V, Jung P, Verdoodt B, Lodygin D,
Epanchintsev A, Menssen A, Meister G and Hermeking H: Differential
regulation of microRNAs by p53 revealed by massively parallel
sequencing: miR-34a is a p53 target that induces apoptosis and
G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fujita Y, Kojima K, Hamada N, Ohhashi R,
Akao Y, Nozawa Y, Deguchi T and Ito M: Effects of miR-34a on cell
growth and chemoresistance in prostate cancer PC3 cells. Biochem
Biophys Res Commun. 377:114–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pogribny IP, Muskhelishvili L, Tryndyak VP
and Beland FA: The tumor-promoting activity of
2-acetylaminofluorene is associated with disruption of the p53
signalIng pathway and the balance between apoptosis and cell
proliferation. Toxicol Appl Pharmacol. 235:305–311. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kojima K, Fujita Y, Nozawa Y, Deguchi T
and Ito M: MiR-34a attenuates paclitaxel-resistance of
hormone-refractory prostate cancer PC3 cells through direct and
indirect mechanisms. Prostate. 70:1501–1512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Luan S, Sun L and Huang F: MicroRNA-34a: A
novel tumor suppressor in p53-mutant glioma cell line U251. Arch
Med Res. 41:67–74. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pramanik D, Campbell NR, Karikari C,
Chivukula R, Kent OA, Mendell JT and Maitra A: Restitution of tumor
suppressor microRNAs using a systemic nanovector inhibits
pancreatic cancer growth in mice. Mol Cancer Ther. 10:1470–1480.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang S, Hao J, Xie F, Hu X, Liu C, Tong
J, Zhou J, Wu J and Shao C: Downregulation of miR-132 by promoter
methylation contributes to pancreatic cancer development.
Carcinogenensis. 32:1183–1189. 2011. View Article : Google Scholar
|
|
54
|
Menghini R, Casagrande V, Cardellini M,
Martelli E, Terrinoni A, Amati F, VasaNicotera M, Ippoliti A,
Novelli G, Melino G, et al: MicroRNA 217 modulates endothelial cell
senescence via silent information regulator 1. Circulation.
120:1524–1532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
vanRooij E: The art of microRNA research.
Circ Res. 108:219–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sarver AL and Subramanian S: Competing
endogenous RNA database. Bioinformation. 8:731–733. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kartha RV and Subramanian S: Competing
endogenous RNAs (ceRNAs): New entrants to the intricacies of gene
regulation. Front Genet. 5:82014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Vasudevan S: Posttranscriptional
upregulation by microRNAs. Wiley Interdiscip Rev RNA. 3:311–330.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lewis BP, Shih IH, JonesRhoades MW, Bartel
DP and Burge CB: Prediction of mammalian microRNA targets. Cell.
115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
ValinezhadOrang A, Safaralizadeh R and
Kazemzadeh-Bavili M: Mechanisms of miRNA-mediated gene regulation
from common downregulation to mRNA-specific upregulation. Int J
Genomics. 2014:9706072014.PubMed/NCBI
|
|
63
|
Fang Y and Nicholl MB: Sirtuin 1 in
malignant transformation: Friend or foe? Cancer Lett. 306:10–14.
2011. View Article : Google Scholar : PubMed/NCBI
|

















