Introduction
The development of cancer vaccines has become a high
priority in the field of cancer treatment. However, there are
numerous parameters that affect the success of an immune response
against an antigen, including the binding of a T-cell receptor to a
major histocompatibility complex (MHC)-bound antigen and antigen
processing. Empirical evaluations of vaccine efficacy parameters
are costly and time-consuming. Thus, bioinformatic approaches may
provide a useful alternative.
The Immune Epitope Database (IEDB) includes
>35,000 human peptides known to either bind to human leukocyte
antigen (HLA) class I or II, or to have other immune receptor
binding properties (1,2). Knowledge of the capacity of a peptide to
bind to antigen-presenting molecules could potentially improve the
selection of cancer vaccine candidates that are based on mutant
peptides, whether these result from cancer drivers or passenger
mutations. In addition, sufficient database development may allow
for a better understanding of any presumed selection against the
binding of cancer peptide neoantigens to MHC molecules as an aspect
of cancer development.
The present study focused on searching for overlaps
of The Cancer Genome Atlas (TCGA) mutant peptides (3,4) and
peptides in the IEDB, in order to discover cancer-related peptides
that have the demonstrable capability to bind to MHC molecules.
Materials and methods
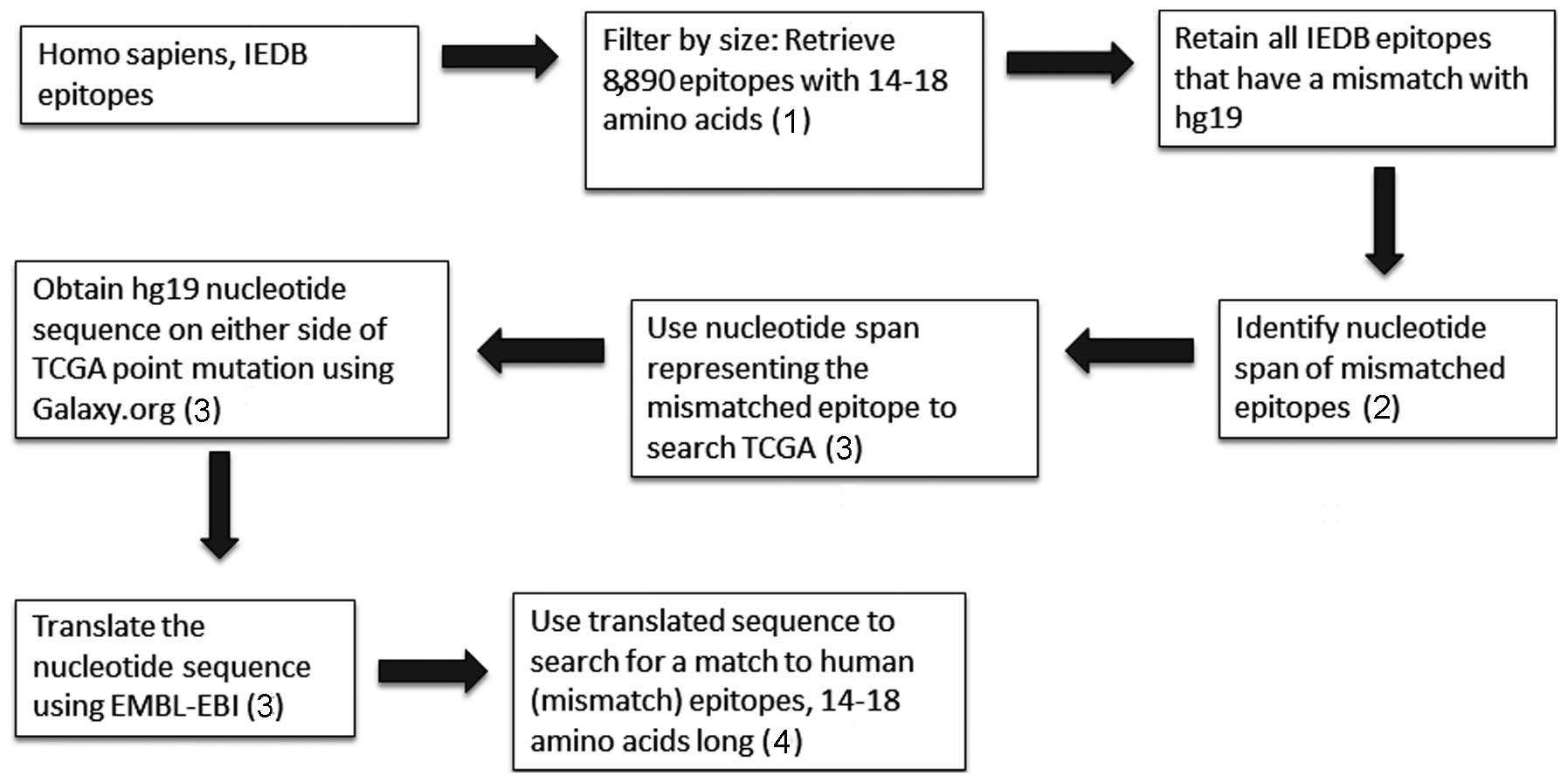
The overview of the approach is provided in Fig. 1. Supporting online material (SOM)
representing each stage of the approach were also used in the
present study (http://www.universityseminarassociates.com/Supporting_online_material_for_scholarly_pubs.php)
(5). Briefly, human epitopes were
downloaded from the IEDB (www.iedb.org)
using the following search terms: Epitope, linear epitope; antigen,
Homo sapiens (human) (ID: 9606, Homo sapiens); host,
humans; assay, all assays; MHC restriction, MHCI and MHCII;
disease, any disease. The results comprised ~35,000 epitopes and
were downloaded as an Excel file. Epitopes that ranged between
14–18 amino acids (AAs) in size were used to determine mismatches
with the human genome version 19 (hg19) reference genome at
genome.ucsc.edu. The nucleotide spans of the
mismatches were obtained by using the BLAT search (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start)
to search a local database, which was created from the TCGA
download portal and consisted of a collection of all TCGA cancer
mutations (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp).
The recovered nucleotide positions were extended with hg19
nucleotides using The Extract Genomic DNA tool at https://usegalaxy.org/. The extended regions were then
translated into all possible reading frames (including forward and
reverse) using the European Molecular Biology Laboratory-European
Bioinformatics Institute to generate a database for screening the
IEDB 14–18 AA set, in order to verify the IEDB matches and to
detect mismatches at the location of the TCGA mutation. All HLA
candidates were removed due to overly extensive sequence variation.
Gene family members that were originally inaccurately regarded as
hg19 mismatches, may be found in the SOM files by Sait et al
(5) (Table
I).
 | Table I.Identification of IEDB peptides that
overlap the position of a mutant amino acid in the TCGA
database. |
Table I.
Identification of IEDB peptides that
overlap the position of a mutant amino acid in the TCGA
database.
| Hugo gene symbol | Normal peptide
sequence (hg19) | IEDB ID number | IEDB peptide (hg19
mismatched amino acid in large type, bold) | hg19 translation of
TCGA mutation on either side of mutation position | TCGA mutation
chromosome number | TCGA cancer
dataset | TCGA nucleotide
number | MHC |
|---|
| ATP1A2 | NPREAKACVVHGSDLK | 103447 | NPRDAKACVVHGSDLK | VVHGSDL | 1 | LUAD/LIHC | 160104960 | HLA-DQB |
|
|
|
|
| ACVVHG | 1 | LIHC | 160104952 |
|
|
|
|
| CVVHGS | 1 | LIHC | 160104955 |
|
|
|
|
| VVHGSDL | 1 | LUAD/LIHC | 160104960 |
| COL2A1 | GEPGIAGFKGEQGPKG | 107398 | GKPGIAGFKGEQGPKG | IAGFKG | 12 | READ |
48380651 | HLA-DRB1 |
| TROVE2 | LQEMPLTALLRNLGKM | 118499 | LQEMPTLALLRNLGKM | ALLRNL | 1 | SKCM | 193045674 | NA |
Results and Discussion
The present study was required to determine whether
detecting an IEDB peptide that had a mismatch at the exact position
of a TCGA mutant AA was possible. Therefore, a search was performed
among the 8,890 IEDB human peptides consisting of 14–18 AAs, with
translated AAs on either side of all TCGA point mutations, to check
for overlap with an IEDB epitope that had a mismatch with the hg19
version of the reference genome. Since the translations represented
exact matches with the hg19 translations, the 8,890 epitopes
consisting of 14–18 AA were searched, allowing for one mismatch
with the translations used, in order to ‘surround’ the location of
the TCGA mutation. According to this protocol, while the TCGA point
mutation-referenced translations overlapped the position of the
TCGA mutation, these translations matched hg19 exactly, thus
requiring the single mismatch standard for searching the
aforementioned 8,890 IEDB epitopes for an exact match.
Numerous IEDB epitopes were identified using this
method; however, following the exclusion of IEDB epitopes that did
not match the gene of the TCGA mutation, only one IEDB peptide had
a non-hg19 AA in the position of the TCGA mutant AA. This IEDB
epitope mapped to integrin subunit β 3 (ITGB3), which is a known
ITGB3 single nucleotide polymorphism. The data supporting this
finding is presented in SOM file no. 5 of Sait et al
(5).
To determine whether the TCGA mutant AA positions
overlapped IEDB peptides that contained a mismatch with the hg19 AA
sequence, without the TCGA position equaling the precise location
of the IEDB mismatched AAs, the protocol indicated in Fig. 1 was followed. The results are provided
in Table I. This protocol indicated
that, following the removal of mismatches attributable to closely
associated family members or mismatches detected anomalously due to
repeats within a protein, 3 IEDB peptides, which were a mismatch to
hg19, also overlapped the position of the TCGA mutant AA. For
details of the results that were obtained by pursuing this
approach, including the discounted IEDB peptides that were
anomalously recovered using the Fig.
1 approach, please see SOM file no. 6 in Sait et al
(5). Overall, these results indicate
that mutant peptides in human cancer overlap apparent mutant
peptides in the IEDB, suggesting that the AAs surrounding TCGA
mutants are not fundamentally a hindrance to MHC binding. Notably,
two of the proteins represented by the overlap of TCGA mutations
and IEDB non-hg19 peptides represent the extracellular matrix,
ITGB3 and collagen type II α 1, an emerging topic in the field of
cancer research (4,6,7).
However, the general paucity of the overlap of the
two databases strongly indicates that, from a bioinformatic
perspective, there is very little information available for
determining which cancer drivers or passenger mutations have the
potential of significant MHC binding. This conclusion is even more
striking considering the extensive MHC polymorphism and protease
activities that could impact binding affinities of cancer peptides
(8).
In conclusion, there is a strong case to be made for
the development of a more comprehensive human immuno-peptidome
project, with the particular aim of determining whether cancer
peptides are selected for the reduced likelihood of MHC
occupancy.
References
|
1
|
He Y and Xiang Z: Databases and in silico
tools for vaccine design. Methods Mol Biol. 993:115–127. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Helmberg W: Bioinformatic databases and
resources in the public domain to aid HLA research. Tissue
Antigens. 80:295–304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akbani R, Ng PK, Werner HM, Shahmoradgoli
M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al: A
pan-cancer proteomic perspective on the cancer genome atlas. Nat
Commun. 5:38872014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parry ML, Ramsamooj M and Blanck G: Big
genes are big mutagen targets: A connection to cancerous, spherical
cells? Cancer Lett. 356:479–482. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sait S, Fawcett T and Blanck G: Supporting
online materials for Overlap of The Cancer Genome Atlas and the
Immune Epitope Database. http://www.universityseminarassociates.com/Supporting_online_material_for_scholarly_pubs.phpAccessed.
June 10–2016
|
|
6
|
Parry ML and Blanck G: Flat cells come
full sphere: Are mutant cytoskeletal-related proteins
oncoprotein-monsters or useful immunogens? Hum Vaccin Immunother.
12:120–123. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naba A, Clauser KR, Whittaker CA, Carr SA,
Tanabe KK and Hynes RO: Extracellular matrix signatures of human
primary metastatic colon cancers and their metastases to liver. BMC
Cancer. 14:5182014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cronin K, Escobar H, Szekeres K,
Reyes-Vargas E, Rockwood AL, Lloyd MC, Delgado JC and Blanck G:
Regulation of HLA-DR peptide occupancy by histone deacetylase
inhibitors. Hum Vaccin Immunother. 9:784–789. 2013. View Article : Google Scholar : PubMed/NCBI
|