Introduction
The use of reversible epidermal growth factor
receptor (EGFR) tyrosine kinase inhibitors (TKIs), including
gefitinib and erlotinib, which function by competitively binding at
the adenosine triphosphate-binding cleft of the receptor kinase
domain of EGFR and consequently blocking the kinase activation and
subsequent downstream signal transduction processes, produces
secondary resistance in the majority of lung cancer patients
(1). Previous studies have
demonstrated that the majority of the acquired resistance is due to
the EGFR T790M mutation (2–6).
Second-generation irreversible EGFR TKIs, including BIBW2992
(afatinib) and PF-002999804, were developed to solve the drug
resistance problem, but did not yield the desired results due to
their narrow therapeutic window (7–9). The
third-generation T790M specific inhibitors, mainly AZD9291 and
CO-1686, are currently under clinical trials in patients with
acquired resistance (7,8).
Formalin-fixed paraffin-embedded (FFPE) samples have
been widely recognized as common clinical materials for EGFR
mutation detection, in which DNA may be partially degraded
(10). EGFR mutation is a
heterogeneous somatic mutation whose abundance may vary widely
(11). Therefore, it is important to
select a highly sensitive detection method for these low quality
DNA samples and for those with low mutation abundances.
The EGFR T790M mutation may originate from
small subclonal populations in the primary tumor, and may become
dominant later on during EGFR-TKIs treatment (12). Early detection of the T790M mutation
is of significance to aid the clinician to adjust the treatment
timely, so that the patient can obtain the most effective therapy
while reducing drug cost and waste. To date, the amplification
refractory mutation system (ARMS) has been widely used in clinical
gene mutation detection, including EGFR, KRAS,
BRAF and phosphatidylinositol-4,5-bisphosphate 3-kinase,
catalytic subunit alpha (PIK3CA), due to its high
sensitivity (13–24).
The cell line H1975 is EGFR T790M-positive
(~50% mutation rate), but in our attempt to develop a T790M
ARMS-based quantitative polymerase chain reaction (qPCR) method,
the cell line H1975 was detected as T790M-positive with low
mutation abundance (~1% mutation rate) using the ARMS-qPCR method
(referred to as the ARMS-qPCR system 1), which was validated as
having a detection limit of 1% in our preliminary study. The
ARMS-qPCR method combines the ARMS primer and a unique fluorescent
probe molecule, with the mutant allele selectively amplified by
ARMS, and the amplified PCR product sensitively and specifically
detected by the fluorescent probe system (24). Subsequent sequencing data revealed
that the cell line H1975 is also a variant homozygous for rs1050171
(also designated as c. G2361A or p. Q787Q). We hypothesized that
rs1050171 may affect the sensitivity of the ARMS method, thereby
producing false-negative results. By searching the National Center
for Biotechnology Information (NCBI) single nucleotide polymorphism
(SNP) database, numerous T790M neighboring SNPs were identified,
which were located within the ARMS primer design range (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=121434569).
The majority of them had no frequency data, with the exception of
rs1050171, which has a high variant allele mutation rate (16.2%) in
the Chinese population. These SNPs may influence the detection
sensitivity of the ARMS-based T790M mutation detection assay, thus
carrying the risk of wrong interpretation.
Since the sensitive detection of the T790M mutation
is very important for the individualized therapy of non-small cell
lung cancer (NSCLC) patients (25),
any factors affecting the sensitivity of the detection method
should be a matter of concern. The present study aimed to
investigate whether rs1050171 affects the detection sensitivity of
the ARMS-based T790M mutation assay, as well as to determine the
frequency of rs1050171 in NSCLC patients, and to identify the
frequency of any other SNPs possibly neighboring T790M in NSCLC
patients.
Materials and methods
Cell line
The H1975 [American Type Culture Collection
(ATCC)® CRL-5908D™] and HT29 (ATCC® HTB-38™)
human tumor cell lines were purchased from the ATCC (Manassas, VA,
USA) in December 2013. The cell lines were grown in standard
conditions and validated by genotyping for EGFR. The
genotypes of the cell lines exactly matched those described in the
Catalog of Somatic Mutations in Cancer (http://cancer.sanger.ac.uk/cosmic/sample/overview?id=1513408).
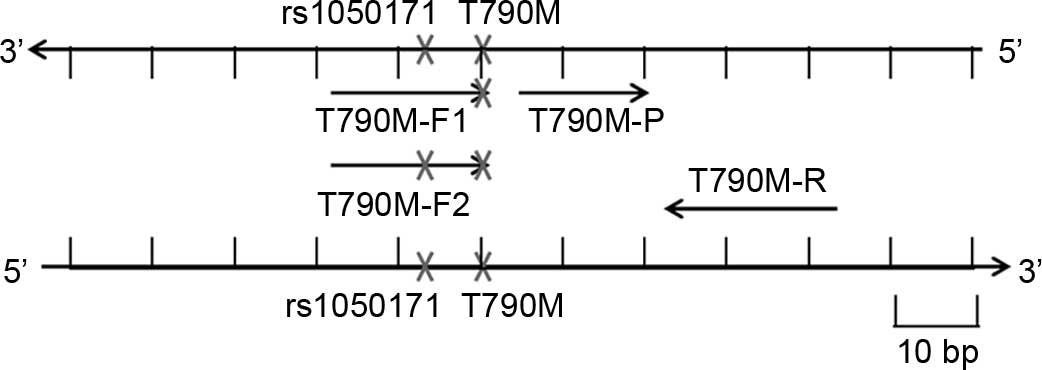
Development of three T790M ARMS-qPCR
systems
The T790M ARMS-qPCR system 1 contained a forward
ARMS primer (named T790M-F1, without rs1050171) designed with a
mismatch at the penultimate nucleotide position of the mutation
site to specifically amplify the T790M mutant allele, a reverse
primer and a minor groove binder probe labeled with the fluorescein
amidite (FAM) fluorescent dye. In the T790M ARMS-qPCR system 2, the
primer T790M-F1 was replaced by T790M-F2 (with rs1050171). In the
T790M ARMS-qPCR system 3, both T790M-F1 and T790M-F2 primers were
included. A reference (RF) reaction system was designed in the
EGFR gene to measure the quantity of both T790M-negative and
T790M mutant alleles, so that the ratio of mutant to wild-type
sequence could be measured. The primers and probes were synthesized
by Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
and Applied Biosystems (Thermo Fisher Scientific, Inc.),
respectively. The 5X PCR buffer, deoxynucleotides and Taq
polymerase were purchased from Takara Bio Inc. (Otsu, Japan). The
primers and probes are listed in Table
I, and the schematic diagram of the three T790M ARMS-PCR
systems is represented in Fig. 1. The
reactions were performed in 25-µl volumes containing 5 µl 5X PCR
buffer, 200 µM dNTPs, 1 µl forward and reverse primers (10 µM
each), 1 µl probe (5 µM), 2 µl template DNA (adjusted to 10 ng/µl)
and 0.1 µl Taq polymerase (5 U/µl). qPCR was conducted using a 7500
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) under the following conditions: Initial denaturation at 95°C
for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1
min, with fluorescence FAM reading at 60°C of each cycle. The
quantification cycle (Cq) represents the threshold at which the
signal was detected above the background fluorescence (25). The ΔCq value was calculated as the
difference between the T790M mutation Cq and the RF Cq.
 | Table I.Primers and probes used in the present
study. |
Table I.
Primers and probes used in the present
study.
| Primers | Sequences | Description |
|---|
| T790M-F1 |
CCTCCTGGCAGCTCATCGT | Forward primer for
the T790M ARMS systems 1 and 3 |
| T790M-F2 |
CCTCCTGGCAACTCATCGT | Forward primer for
the T790M ARMS systems 2 and 3 |
| T790M-R |
TGTTCCCGGACATAGTCCAGG | Reverse primer for
the three T790M ARMS systems |
| RF-F |
GATCCCAGAAGGTGAGAAAGTTAA | Forward primer for
the RF system |
| RF-R |
CACAGCAAAGCAGAAACTCACA | Forward primer for
the RF system |
|
| Probes | Sequences | Description |
|
| T790M-P | CTCATGCCCTTCGGCT | Probe for the three
T790M ARMS systems |
| RF-P | AAGCCAACAAGGAAAT | Probe for the RF
system |
Comparison of the three ARMS-qPCR
systems to detect T790M mutation in mixed samples with or without
rs1050171 and in the cell line H1975
The T790M mutant plasmid 1 (without rs1050171) was
developed and quantified according to Zhang et al (25). The T790M mutant plasmid 2 (with
rs1050171) was developed as follows: A DNA fragment containing the
T790M mutation and rs1050171 was amplified from the genomic DNA
(gDNA) of the cell line H1975 using primers T790M-F2 and T790M-R.
The PCR cycling conditions were 94°C for 5 min; followed by 40
cycles at 94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min; and
a final extension at 72°C for 10 min. The PCR product was then
ligated into the pEASY-T1 vector (Beijing TransGen Biotech Co.,
Ltd., Beijing, China), and DNA sequencing was used to verify the
accuracy of the fragment. Different amounts of T790M mutant
plasmids 1 or 2 were mixed with 20 ng wild-type EGFR HT29
gDNA to yield 1, 2, 5, 10, 20 and 50% mutation rates, respectively.
The two series mixed samples and the gDNA of the cell line H1975
were used to explore the sensitivity of the three ARMS-qPCR
systems, and the detection results were compared to assess the
influence of rs1050171 on the effective detection of T790M.
FFPE tumor samples from NSCLC
patients
A total of 670 FFPE tumor samples (numbered as
N001-N670) obtained from NSCLC patients between January 2013 and
December 2014 were collected from Tongji Hospital (Wuhan, China),
of which, 419 (62.6%) cases were males and 250 (37.4%) cases were
females. The quantity of tumor cells in all samples was >50%, as
confirmed by two skilled pathologists. The study was approved by
Clinical Trial Ethics Committee of Huazhong University of Science
and Technology (Wuhan, China), and written informed consent was
obtained from all patients. gDNA was extracted using the QIAamp DNA
FFPE Tissue kit (cat. no. 56404; Qiagen GmbH, Hilden, Germany), and
was quantified by the measurement of optical density (OD) at 260 nm
using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). The gDNA
purity was evaluated by the measurement of the OD260/OD280 ratio
(all gDNA samples measured between 1.90 and 1.98). The T790M
mutation statuses of all the 670 FFPE samples were detected by
sequencing and with the ARMS-qPCR system 1. For samples with T790M
mutation detected by the ARMS-qPCR system 1, or for samples with
T790M amplification but whose ΔCq values were above the cut-off
value of 7 and were homozygous mutant genotype of rs1050171, the
ARMS-qPCR system 3 was used instead to detect the T790M
mutation.
EGFR exon 20 sequencing
EGFR exon 20 sequencing was performed by
Beijing Genomics Institute (Shenzhen, China), and the results were
analyzed by alignment with the wild-type sequence. The T790M 30-bp
5′ near sequence and the 30-bp 3′ near sequence were also analyzed
to evaluate the presence of potential SNPs, whose frequencies were
also calculated.
Results
T790M mutation detection results of
the cell line H1975 and the mixed plasmid samples by the three
ARMS-qPCR systems
With a detection limit of 1%, the positive results
for the three ARMS-qPCR systems were all defined as having a Cq
<36 and a ΔCq lower than the cut-off value of 7.5. According to
the sequencing results, the cell line H1975 appears to have the
T790M mutation (~50% mutation rate) and has a homozygous mutant
genotype of rs1050171 (Fig. 2A). The
cell line was detected as T790M-positive but with low mutation
abundance (~1% mutation rate) by the ARMS-qPCR system 1, and was
effectively detected as definite T790M mutation (~50% mutation
rate) by the ARMS-qPCR systems 2 and 3, which contain a forward
ARMS primer with rs1050171 (Fig.
2B).
For the two series mixed samples, the ARMS-qPCR
system 1 detected samples with 1% T790M mutant plasmid 1 (without
rs1050171) and 50% T790M mutant plasmid 2 (with rs1050171) in a
background of 20 ng HT29 gDNA (Fig. 2C
and D). The ARMS-qPCR system 2 detected templates containing
20–50% T790M mutant plasmid 1 and 1% T790M mutant plasmid 2
(Fig. 2E and F). For the ARMS-qPCR
system 3, samples with as low as 1% T790M mutant plasmids 1 or 2
could be effectively detected (Fig. 2G
and H). Since the ARMS-qPCR system 2 exhibited problems in
detecting samples with T790M mutation but without rs1050171, this
system was not further studied.
Sequencing results of the 670 FFPE
samples
For SNP analysis, rs1050171 was detected in numerous
cases, and the frequency of the variant-A allele was ≤28.2%, with
wild-type genotype accounting for 47.2%, heterozygous genotype for
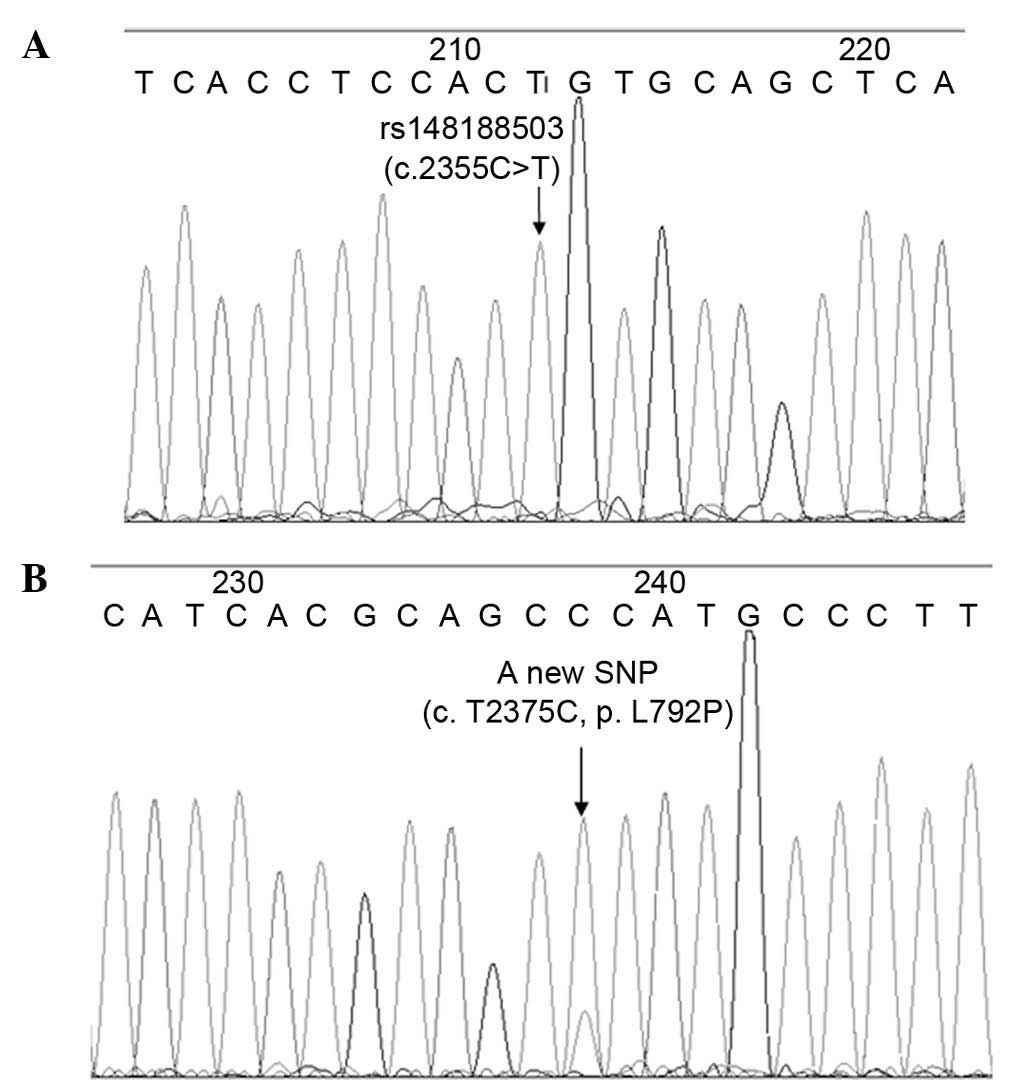
49.2% and variant homozygous genotype for 3.6%. The sample N558 was
observed to be variant homozygous for rs148188503 (c. C2355T),
which is a synonymous mutation with a mutation rate of 0.15%
(Fig. 3A). In the sample N310, a
novel SNP with a base substitution (c. T2375C) in the EGFR
gene (exon 20, position 792, p. L792P) was observed, which was not
included in the SNP database of NCBI (Fig. 3B). In total, 5 cases (0.75%) appeared
to have the T790M mutation. Detailed information regarding the
mutation frequencies of T790M neighboring SNPs in the 670 FFPE
samples analyzed is presented in Table
II.
 | Table II.Mutation frequencies of T790M
neighboring SNPs in 670 formalin-fixed paraffin-embedded
samples. |
Table II.
Mutation frequencies of T790M
neighboring SNPs in 670 formalin-fixed paraffin-embedded
samples.
|
| rs1050171 | rs148188503 | c. T2375C, p.
L792P |
|---|
|
|
|
|
|
|---|
| SNPs | G/G | G/A | A/A | C/C | C/T | T/T | T/T | T/C | C/C |
|---|
| Cases, n | 316 | 330 | 24 | 669 | 0 | 1 | 669 | 1 | 0 |
| (%) | (47.20) | (49.20) | (3.60) | (99.85) | (0.00) | (0.15) | (99.85) | (0.15) | (0.00) |
T790M mutation detection results by
the different ARMS-qPCR systems
Of the 670 cases, 6 cases (0.9%) appeared to have
the T790M mutation by the T790M ARMS-qPCR system 1. Sample N067,
which was firstly determined as T790M-negative by sequencing, it
was identified to be T790M-positive by the ARMS-qPCR system 1. Of
the 19 samples with T790M mutation, or with T790M amplification but
with ΔCq values above the cut-off value of 7.5 and which were
homozygous mutant genotype of rs1050171, the T790M ARMS-qPCR system
3 was used to detect the T790M mutation. As expected, sample N094
was determined to have the T790M mutation (Fig. 4).
Comparison of the T790M mutation
detection results by different methods and the corresponding
rs1050171 mutation analysis
Of the 670 FFPE samples analyzed, 5 cases were
detected as having the T790M mutation by sequencing, and 6 cases
were identified as having the T790M mutation by the ARMS-qPCR
system 1 method. The consistency of the two methods was 99.85%
(669/670). Sample N094, which is variant homozygous for rs1050171,
was firstly detected as T790M-negative by the T790M ARMS-qPCR
system 1, but was confirmed to be T790M-positive by the T790M
ARMS-qPCR system 3. Of the 7 FFPE samples with the T790M mutation,
3, 3 and 1 cases were wild-type, heterozygous mutant and homozygous
mutant genotypes of rs1050171, respectively. Detailed results of
these analyses are presented in Table
III.
 | Table III.Detection results of various samples
with the T790M mutation using different methods. |
Table III.
Detection results of various samples
with the T790M mutation using different methods.
|
|
|
| Sequencing |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Sample name | Gender | Pathology | rs1050171 | T790M | T790M by the
ARMS-qPCR system 1 | T790M by the
ARMS-qPCR system 3 |
|---|
| N019 | Female | ADC | G/G | Positive | Positive | Positive |
| N060 | Female | ADC | G/G | Positive | Positive | Positive |
| N067 | Female | ADC | G/A | Negative | Positive | Positive |
| N072 | Female | ADC | G/G | Positive | Positive | Positive |
| N333 | Male | ADC | G/A | Positive | Positive | Positive |
| N512 | Male | ADC | G/A | Positive | Positive | Positive |
| N094 | Female | ADC | A/A | Negative | Negative | Positive |
Discussion
Three T790M ARMS-qPCR systems were developed in the
present study: i) The ARMS-qPCR system 1 (without rs1050171 in the
forward ARMS primer), which effectively detected samples with as
low as 1% T790M mutant plasmid 1 (without rs1050171) and with 50%
T790M mutant plasmid 2 (with rs1050171); ii) the ARMS-qPCR system 2
(with rs1050171 in the forward ARMS primer), which detected samples
with 20–50% T790M mutant plasmid 1 and with 1% T790M mutant plasmid
2; and iii) the ARMS-qPCR system 3 (where the two forward ARMS
primers mentioned above were included), which effectively detected
samples with as low as 1% T790M mutant plasmids 1 or 2. For the
cell line H1975, the ARMS-qPCR system 1 detected the T790M mutation
with low abundance (~1% mutation rate), and the ARMS-qPCR systems 2
and 3 effectively detected it with high abundance (~50% mutation
rate, consistent with the sequencing results). These results
preliminarily indicated that the existence of rs1050171 affects the
sensitivity of the ARMS-qPCR system used in the present study. To
further confirm this result, 670 FFPE samples from NSCLC patients
were used to detect the T790M mutation by sequencing and by the
ARMS-qPCR system 1, and partial samples were detected by the
ARMS-qPCR system 3. In the 670 FFPE samples, the variant-A allele
frequency of rs1050171 was 28.2%, with the wild-type, heterozygous
and homozygous mutant genotype accounting for 47.2, 49.2 and 3.6%,
respectively. In addition, the results of 669 cases obtained by
sequencing and by the ARMS-qPCR system 1 were consistent (99.85%).
Sample N067, which was firstly identified as T790M-negative and
rs1050171 wild-type by sequencing, was identified as exhibiting the
T790M mutation by the ARMS-qPCR systems 1 and 3, thus illustrating
the higher sensitivity of the ARMS-qPCR method compared with direct
sequencing. Notably, sample N094, which was detected as
T790M-negative by sequencing and by the ARMS-qPCR system 1, was
determined to have the T790M mutation by the ARMS-qPCR system 3.
These results illustrate that rs1050171 actually influences the
sensitivity of the ARMS-based T790M mutation detection method,
particularly in samples with low abundance of T790M mutation.
A large number of patients who are resistant to
EGFR-TKIs often harbor a pre-existing T790M EGFR mutation at very
low levels within the original tumor population, which leads to
resistance following treatment (12).
Screening patients for low levels of T790M EGFR mutation prior to
administering EGFR-TKIs treatment may be useful to assess the
possibility of disease relapse. Furthermore, monitoring the
presence of T790M mutation in plasma during EGFR-TKIs treatment may
be useful for future clinical decision making. In order to detect
these low-level T790M variations in the primary tumor or their
progression in plasma, it is important to apply reliable and
sensitive mutation detection methods (12). With a detection limit of 1%, the ARMS
method is a sensitive, convenient and economic approach that is
widely used in point mutation detection, and any factor affecting
the sensitivity of this method should be addressed (24). According to the SNP database of NCBI,
rs1050171 is an SNP with high mutation frequency located 8 bp prior
to T790M. Under the influence of rs1050171, the T790M mutation in
the gDNA of the cell line H1975 and in the FFPE sample N094 could
not be effectively detected in the present study. Of the 670 FFPE
samples analyzed, 6 and 7 cases were identified to have the T790M
mutation by the ARMS-qPCR system 1 and 3, respectively, and the
false-negative rate (1/7) was 14.3%. Considering the significance
of early detection of low-abundance T790M mutation, the
false-negative rate resulted from rs1050171 should not be
underestimated.
Among the 7 FFPE samples with the T790M mutation, 3
(3/7, 42.9%), 3 (3/7, 42.9%) and 1 cases (1/7, 14.3%) were
wild-type, heterozygous and homozygous mutant genotype of
rs1050171, respectively. Of the 316 wild-type, 330 heterozygous and
24 variant homozygous cases for rs1050171, 3 (3/316, 0.95%), 3
(3/330, 0.91%) and 1 (1/24, 4.17%) cases had the T790M mutation,
respectively. The T790M mutation rate between the wild-type and the
heterozygous genotype of rs1050171, and the frequencies of the
wild-type and heterozygous genotypes of rs1050171 in samples with
the T790M mutation, are consistent with those of the total cases.
The deviation of samples that were variant homozygous for rs1050171
may result from the inadequate cases, since there was only one
sample with the T790M mutation that was variant homozygous for
rs1050171. No obvious association appears to exist between the
mutation frequencies of T790M and rs1050171.
As for the influence of other two SNPs (rs148188503
and c. T2375C, p. L792P) on the ARMS-based T790M mutation assay,
rs148188503 was observed to be located 14 bp prior to T790M, which
was farther away from T790M and had a lower mutation rate (0.15%)
compared with rs1050171. The novel SNP (c. T2375C, p. L792P) is
located 6 bp beyond T790M, which is in the design range of the
reverse ARMS primer, and has a low mutation rate (0.15%). Since no
high frequency SNPs located beyond T790M were detected in the
present study, another possible solution is designing ARMS primers
according to the other strand of the template. Further
investigation about the influence of these two SNPs on ARMS-based
T790M mutation assays is still required.
According to the ARMS principle, the last 3 bp in
the 3′ end of the ARMS primers are very important for the correct
recognition and binding of the primer to the template (26–28). As a
general rule, the mismatched bases are often designed in the last 3
bp of the ARMS primer to discriminate between the wild-type and the
mutant alleles (26–28). Notably, in the present study, the
rs1050171 was observed to be located in the eighth base from the 3′
end of the ARMS primer, and the results revealed that it really
affected the sensitivity of T790M mutation detection, particularly
in samples with low mutation rates. It is possible that a similar
situation may occur in other EGFR mutations and in point
mutation in other genes, including KRAS c.34G>T (G12C),
BRAF c.1799T>A (V600E) and PIK3CA c.1633G>A
(E545K). The present study highlighted the risk associated with the
target neighboring SNPs (as far as 8 bp away from T790M mutation
site in the current study), which may influence the effective
detection of the target site, and should be considered when
detecting novel point mutations.
In conclusion, the existence of T790M neighboring
rs1050171 (located at 8 bp prior to T790M) reduces the sensitivity
of the ARMS-based T790M mutation detection assay and produces a
14.3% false-negative rate. The influence of target neighboring SNPs
on the effective detection of the target mutation must be taken
into consideration when starting a novel point mutation detection
project.
Acknowledgements
The authors would like to thank Dr Congli Cai, Mr.
Zhe Zhang and Mrs. Liqiong Li, researchers at Wuhan YZY Medical
Science and Technology Co. Ltd. (Wuhan, China), for their advice
and suggestions in developing the ARMS-based T790M mutation assay
and for the construction of the plasmid samples.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
EGFR T790M
|
epidermal growth factor receptor gene
T790M mutation
|
|
ARMS
|
amplification refractory mutation
system
|
|
SNP
|
single nucleotide polymorphism
|
References
|
1
|
Wang J, Ramakrishnan R, Tang Z, Fan W,
Kluge A, Dowlati A, Jones RC and Ma PC: Quantifying EGFR
alterations in the lung cancer genome with nanofluidic digital PCR
arrays. Clini Chem. 56:623–632. 2010. View Article : Google Scholar
|
|
2
|
Kim Y, Ko J, Cui Z, Abolhoda A, Ahn JS, Ou
SH, Ahn MJ and Park K: The EGFR T790M mutation in acquired
resistance to an irreversible second-generation EGFR inhibitor. Mol
Cancer Ther. 11:784–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kuang Y, Rogers A, Yeap BY, Wang L,
Makrigiorgos M, Vetrand K, Thiede S, Distel RJ and Jänne PA:
Noninvasive detection of EGFR T790M in gefitinib or erlotinib
resistant non-small cell lung cancer. Clinical cancer research: An
official journal of the American Association for Cancer Research.
15:2630–2636. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun JM, Ahn MJ, Choi YL, Ahn JS and Park
K: Clinical implications of T790M mutation in patients with
acquired resistance to EGFR tyrosine kinase inhibitors. Lung
Cancer. 82:294–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamada T, Azuma K, Muta E, Kim J, Sugawara
S, Zhang GL, Matsueda S, Kasama-Kawaguchi Y, Yamashita Y, Yamashita
T, et al: EGFR T790M mutation as a possible target for
immunotherapy; identification of HLA-A*0201-restricted T cell
epitopes derived from the EGFR T790M mutation. PloS One.
8:e783892013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Denis MG, Vallée A and Théoleyre S: EGFR
T790M resistance mutation in non small-cell lung carcinoma. Clin
Chim Acta. 444:81–85. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin L and Bivona TG: Mechanisms of
resistance to epidermal growth factor receptor inhibitors and novel
therapeutic strategies to overcome resistance in NSCLC patients.
Chemother Res Pract. 2012:8172972012.PubMed/NCBI
|
|
9
|
Zhou W, Ercan D, Chen L, Yun CH, Li D,
Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, et al:
Novel mutant-selective EGFR kinase inhibitors against EGFR T790M.
Nature. 462:1070–1074. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yung TK, Chan KC, Mok TS, Tong J, To KF
and Lo YM: Single-molecule detection of epidermal growth factor
receptor mutations in plasma by microfluidics digital PCR in
non-small cell lung cancer patients. Clin Cancer Res. 15:2076–2084.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thomas RK, Nickerson E, Simons JF, Jänne
PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, et
al: Sensitive mutation detection in heterogeneous cancer specimens
by massively parallel picoliter reactor sequencing. Nat Med.
12:852–855. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guha M, Castellanos-Rizaldos E and
Makrigiorgos GM: DISSECT method using PNA-LNA clamp improves
detection of T790M mutation. PloS One. 8:e677822013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bai H, Wang Z, Wang Y, Zhuo M, Zhou Q,
Duan J, Yang L, Wu M, An T, Zhao J and Wang J: Detection and
clinical significance of intratumoral EGFR mutational heterogeneity
in Chinese patients with advanced non-small cell lung cancer. PloS
One. 8:e541702013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Board RE, Ellison G, Orr MC, Kemsley KR,
McWalter G, Blockley LY, Dearden SP, Morris C, Ranson M, Cantarini
MV, et al: Detection of BRAF mutations in the tumour and serum of
patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma
phase II study. Br J Cancer. 101:1724–1730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chu H, Zhong C, Xue G, Liang X, Wang J,
Liu Y, Zhao S, Zhou Q and Bi J: Direct sequencing and amplification
refractory mutation system for epidermal growth factor receptor
mutations in patients with non-small cell lung cancer. Oncol Rep.
30:2311–2315. 2013.PubMed/NCBI
|
|
16
|
Ellison G, Donald E, McWalter G, Knight L,
Fletcher L, Sherwood J, Cantarini M, Orr M and Speake G: A
comparison of ARMS and DNA sequencing for mutation analysis in
clinical biopsy samples. J Exp Clin Cancer Res. 29:1322010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franklin WA, Haney J, Sugita M, Bemis L,
Jimeno A and Messersmith WA: KRAS mutation: Comparison of testing
methods and tissue sampling techniques in colon cancer. J Mol
Diagn. 12:43–50. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fukuoka M, Wu YL, Thongprasert S,
Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu
DT, Saijo N, et al: Biomarker analyses and final overall survival
results from a phase III, randomized, open-label, first-line study
of gefitinib versus carboplatin/paclitaxel in clinically selected
patients with advanced non-small-cell lung cancer in Asia (IPASS).
J Clin Oncol. 29:2866–2874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hamfjord J, Stangeland AM, Skrede ML,
Tveit KM, Ikdahl T and Kure EH: Wobble-enhanced ARMS method for
detection of KRAS and BRAF mutations. Diagn Mol Pathol. 20:158–165.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harlé A, Lion M, Lozano N, Husson M,
Harter V, Genin P and Merlin JL: Analysis of PIK3CA exon 9 and 20
mutations in breast cancers using PCR-HRM and PCR-ARMS: Correlation
with clinicopathological criteria. Oncol Rep. 29:1043–1052.
2013.PubMed/NCBI
|
|
21
|
Huang T, Zhuge J and Zhang WW: Sensitive
detection of BRAF V600E mutation by amplification refractory
mutation system (ARMS)-PCR. Biomark Res. 1:32013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Liu B, Li XY, Li JJ, Qin HF, Tang
CH, Guo WF, Hu HX, Li S, Chen CJ, et al: A comparison of ARMS and
direct sequencing for EGFR mutation analysis and tyrosine kinase
inhibitors treatment prediction in body fluid samples of
non-small-cell lung cancer patients. J Exp Clin Cancer Res.
30:1112011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Machnicki MM, Glodkowska-Mrowka E,
Lewandowski T, Ploski R, Wlodarski P and Stoklosa T: ARMS-PCR for
detection of BRAF V600E hotspot mutation in comparison with
real-time PCR-based techniques. Acta Biochim Pol. 60:57–64.
2013.PubMed/NCBI
|
|
24
|
Ogasawara N, Bando H, Kawamoto Y, Yoshino
T, Tsuchihara K, Ohtsu A and Esumi H: Feasibility and robustness of
amplification refractory mutation system (ARMS)-based KRAS testing
using clinically available formalin-fixed, paraffin-embedded
samples of colorectal cancers. Jpn J Clin Oncol. 41:52–56. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang B, Xu CW, Shao Y, Wang HT, Wu YF,
Song YY, Li XB, Zhang Z, Wang WJ, Li LQ and Cai CL: Comparison of
droplet digital PCR and conventional quantitative PCR for measuring
gene mutation. Exp Ther Med. 9:1383–1388. 2015.PubMed/NCBI
|
|
26
|
Pettersson M, Bylund M and Alderborn A:
Molecular haplotype determination using allele-specific PCR and
pyrosequencing technology. Genomics. 82:390–396. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vlassov VV, Laktionov PP and Rykova EY:
Circulating nucleic acids as a potential source for cancer
biomarkers. Curr Mol Med. 10:142–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye S, Dhillon S, Ke X, Collins AR and Day
IN: An efficient procedure for genotyping single nucleotide
polymorphisms. Nucleic Acids Res. 29:E88. 2001. View Article : Google Scholar : PubMed/NCBI
|