Introduction
Lung cancer is the most frequently diagnosed cancer
and the leading cause of cancer-related mortality worldwide in both
males and females (1), accounting for
more than 1.4 million mortalities per year. Moreover, with the high
and increasing number of smokers in a number of Asian and
developing countries, it is anticipated that the incidence of lung
cancer will continue to increase over the next decades. The rates
of lung cancer are mainly determined by smoking patterns, although
other factors, including medical, occupational and environmental
radiation exposure, have also been demonstrated to increase the
risk of lung cancer (2).
Lung cancer is a heterogeneous disease that has
historically been divided into two main types based on the disease
patterns and treatment strategies: small cell-lung cancer (SCLC)
and non-small cell-lung cancer (NSCLC) (3,4). The
expressions of various oncogenes have been investigated in NSCLC
and SCLC. The complex multiple genetic lesions in lung cancer are
composed of two major forms (5):
mutations activating the dominant cellular proto-oncogenes, and as
those inactivating the tumor suppressor genes. The activation of
RAS, MYC and HER-2/NEU genes is usually
associated with an adverse prognosis of lung cancer (6–8), while the
overexpression of tumor suppressor genes including VHL,
RB and p53 demonstrate a positive outcome in the
recovery of lung cancer patients (2).
Chemotherapy remains the cornerstone of treatment
for limited- and extensive-stage lung cancer. Surgery is also
considered for certain stages (9).
However, these traditional treatments only provide a modest benefit
with considerable toxicity for advanced lung cancer (10). There is an urgent requirement to
develop alternative therapies to enable the monitoring of lung
cancer treatment efficacy and assessment of the risk of metastases.
Advances in our understanding of molecular genetics in lung cancer
have led to the identification of key genetic aberrations in lung
cancer. The potential of certain clinically relevant driver
molecular lesions, including epidermal growth factor receptor
(EFGR) mutations, ROS1 translocation and BRAF
mutations, has already been confirmed (11).
In the present study, we focused on the LIM and SH3
protein 1 (LASP-1), another key molecule involved in development of
multiple cancers, and attempted to elucidate its effect on the
oncogenesis of lung cancer. The gene encodes a membrane-bound
protein containing 261 amino acids with an N-terminal LIM domain
(12,13). The exact function of LASP-1 in lung
cancer is still not well understood; however, its expression is
noted to be increased in a number of malignant tumors, including
breast, colorectal, hepatocellular and bladder cancer (14–18). For
the first time, we determined the expression level of LASP-1 in
lung cancer using reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and western blot analysis. We also studied
the potential function of LASP-1 in lung cancer cell growth,
apoptosis and migration by small interfering RNA (siRNA)
transfection. The aim of our study was to evaluate the function of
LASP-1 in lung cancer and clarify the prognostic value of LASP-1 in
the clinic.
Materials and methods
Patients and sample collection
Twenty patients were enrolled in the current study.
Sections were obtained from tumor tissues and para-carcinoma
tissues, and examined independently by senior pathologists. Parts
of each sample were frozen immediately following surgery and stored
at −135°C for RNA and protein extraction. The human lung cancer
cell line A549 was purchased from ATCC (Manassas, VA, USA). The
ethics committee of the Cancer Hospital of Shantou University
Medical College approved the screening, inspection and data
collection of the patients, and all subjects signed a written
informed consent form. The study was undertaken in accordance with
the provisions of the Declaration of Helsinki.
RNA isolation and RT-qPCR
LASP-1 mRNA expression in cancer tissues was
analyzed by one-step RT-qPCR using an RNA Master SYBR-Green I kit
(Roche Applied Science, Indianapolis, IN, USA). The whole RNA of
the lung tissue of each sample was extracted with TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions. cDNA was synthesized with the
PrimeScript RT reagent kit (Takara Biotechnology Co. Ltd., Dalian,
China). RT-qPCR was then performed using 2X SYBR-Green PCR master
mix (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) in an ABI PRISM 7500 system. The relative level of LASP-1
mRNA was normalized to that of β-actin and analyzed by the
comparative Ct method.
Western blot analysis
The protein products of LASP-1 and the reference
GAPDH were extracted from the lung tissue of the patients. The
extracts were boiled using loading buffer for 5 min and then
subjected to 10% sodium dodecylsulfate polyacrylamide gel
electrophoresis. The protein products were then transferred onto
polyvinylidene difluoride sheets. The membranes were washed with
Tris-buffered saline and Tween-20 for 20 min and the procedure was
repeated three times. Then the membranes were incubated with
antibody overnight at room temperature. After three more washes,
secondary antibody was added and the membrane was incubated for
another 4 hours. The blots were developed using Beyo ECL Plus
reagent (Beyotime Institute of Biotechnology, Haimen, China) after
three final washes. The results of analysis were determined in the
Gel Imaging system (Liuyi Factory, Beijing, China). The expression
levels were calculated with Quantity One software (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
siRNA transfection
The target sequence for LASP-1-specific siRNA
(5′-CTGGATAAGTTCTGGCATA-3′) and the negative control (NC) siRNA
were obtained from Genechem Biotech (Shanghai, China). The human
lung cancer cell line A549 was subjected to one of three
treatments: no treatment (control group), treatment with NC-siRNA,
and treatment with LASP-1-siRNA. Transfections were
conducted using Lipofectamine 2000 reagent (Invitrogen Life
Technologies) following the manufacturer's instructions, and
subsequent selection was conducted using 400 µg/ml puromycin
(Amresco, Solon, OH, USA). The protein products of LASP-1 and
reference protein GAPDH were extracted from the cells, and western
blot analysis was conducted as described above.
Cell proliferation
The cell proliferation ability of the different
treatments was analyzed using a WST-8 Cell Counting kit-8 (Beyotime
Institute of Biotechnology). Briefly, 50 µl exponentially growing
cells (2×105 cells/ml) in RPMI-1640 medium (100 µl)
containing 10% fetal bovine serum were seeded into a 96-well plate.
For each group, cells were incubated for 0, 24, 48, 72 and 96 h,
respectively, and each time point was represented by three
replicates. Following incubation, CCK-8 solution (10 µl) was added
to each well and the cultures were incubated at 37°C for 90 min.
The optical density values in the different wells were recorded
using a microplate reader (Rayto Life and Analytical Science Co.,
Ltd, Shenzhen, China) at 450 nm.
Cell apoptosis
Cells with different treatments were seeded at
2×105 per 35-mm culture dish for 48 h. Apoptosis was
assessed by Annexin V-Cy5 and propidium iodide (PI) staining
(K103-25; BioVision, Milpitas, CA, USA) followed by
fluorescence-activated cell sorting (FACS) analysis. The cells were
pelleted and resuspended in Annexin V binding buffer (10 mM HEPES,
150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM
CaCl2; pH 7.4) containing Annexin V-Cy5 and 1 mg/ml PI.
Following incubation at room temperature for 5 min, the cells were
analyzed with a FACSCalibur flow cytometer (BD Biosciences, San
Jose, CA, USA). The total apoptotic cell percentage of all cells
was determined as UR + LR, which is the sum of the later apoptosis
rate (UR, upper-right quadrant, showing advanced stage apoptosis
cell percentage) and the early apoptosis rate (LR, lower-right
quadrant, showing the prophase apoptosis cell percentage).
Cell migration
Cells with different treatments were incubated for
48 h and starved overnight, then 100 µl incubation medium (with 1
mM MgCl2) containing 1×105 cells was seeded
in the upper chamber of bovine serum albumin-coated 8-µM pore size
Transwell chambers (Corning, Cambridge, MA, USA). Cells were
allowed to migrate through the porous membrane for 4 h at 37°C.
Cells remaining on the upper surface were removed with a cotton
carrier. The lower surfaces of the membranes were stained in a
solution of 1% (w/v) crystal violet in 2% ethanol for 30 sec and
then rinsed in distilled water. Cell-associated crystal violet was
extracted by incubation in 10% acetic acid for 20 min. Non-migrated
cells were removed with cotton swabs. Cell images were obtained
under a phase-contrast microscope (Olympus, Tokyo, Japan).
Statistical analysis
All the data were expressed as the means ± standard
deviation. Multiple comparisons were conducted using the least
significant difference method; P<0.05 was considered to indicate
a statistically significant difference. All statistical analyses
were conducted using SPSS version 19.0 (IBM SPSS, Armonk, NY,
USA).
Results
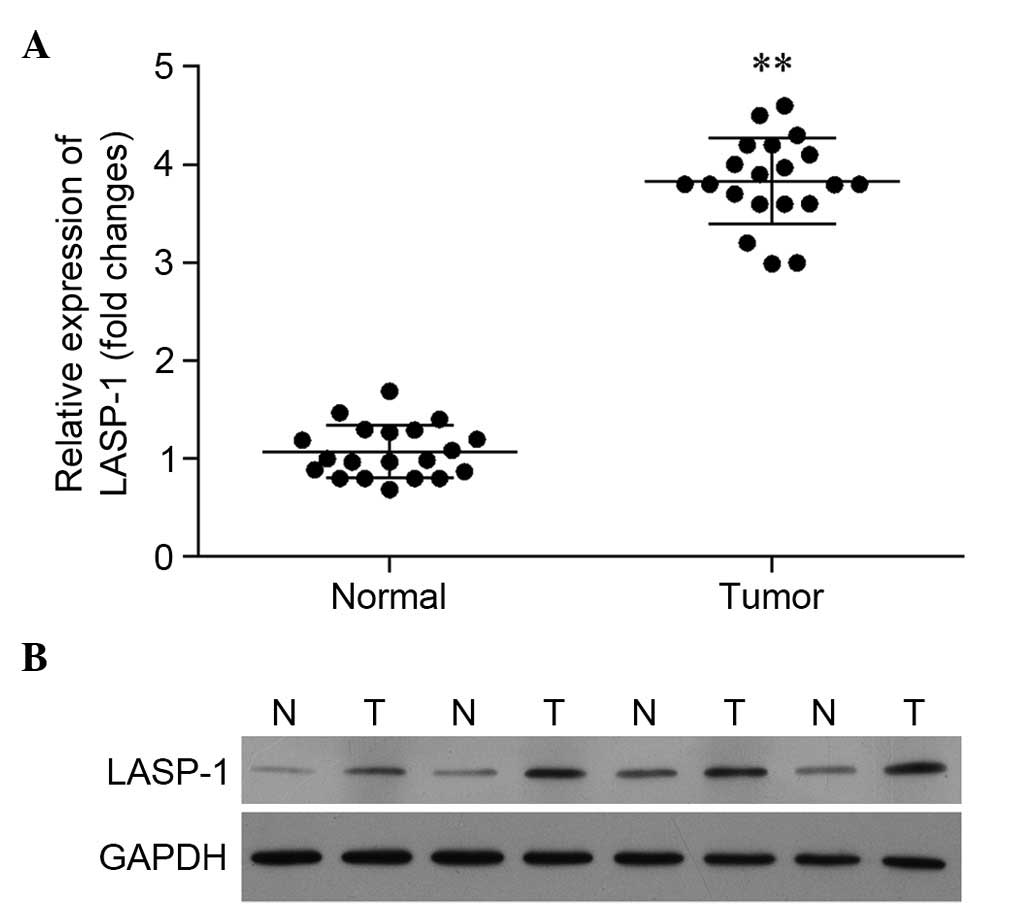
Expression of LASP-1 is significantly
upregulated in tumor tissues
Based on the results of RT-qPCR, the relative
expression of the LASP-1 gene in tumor tissues was
significantly higher than that in para-carcinoma tissues (Fig. 1A). A similar pattern was observed in
western blot analysis. In Fig. 1B,
the expression of LASP-1 in four patients is illustrated, and the
results reveal significantly enhanced synthesis of LASP-1 in the
tumor tissues.
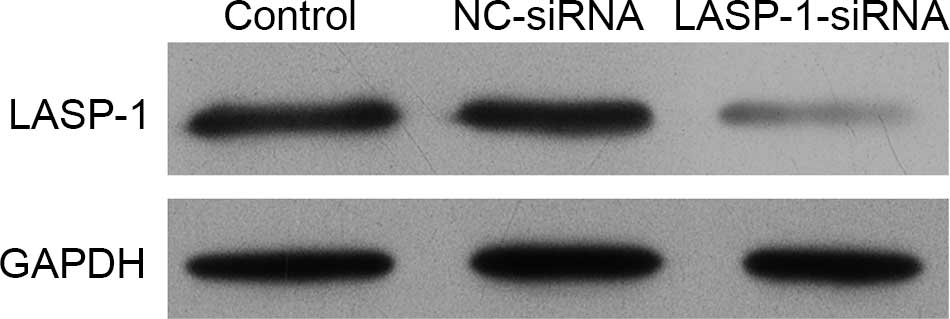
siRNA transfection of LASP-1 gene
blocks the synthesis of LASP-1
The human lung cancer cell line A549 was subjected
to one of three different treatments and then the synthesis of
LASP-1 was detected using western blot analysis. It was
demonstrated that the stable transfection of LASP-1-siRNA
significantly blocked the synthesis of LASP-1 compared with the
other two treatments (Fig. 2).
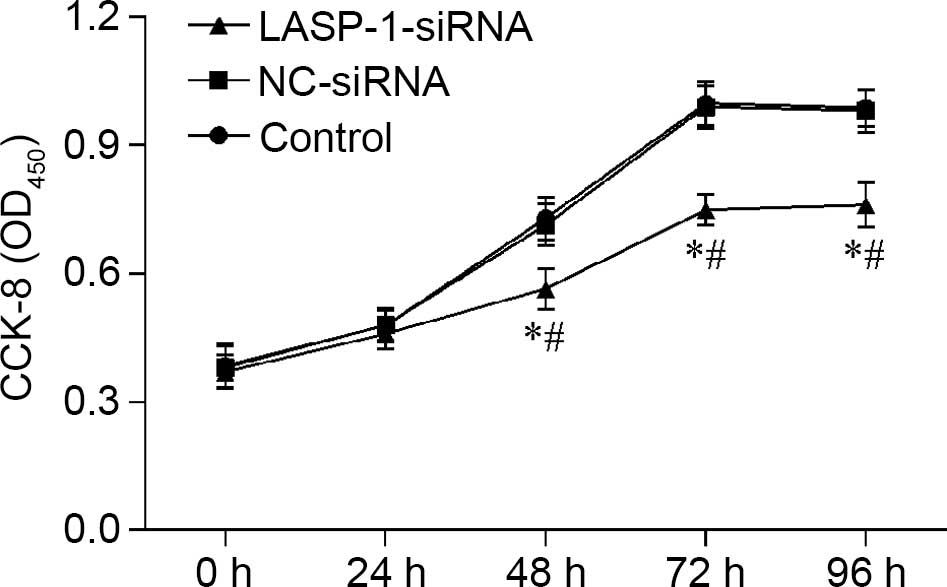
Downregulation of LASP-1 influences
the proliferation, apoptosis and migration of A549 cell line
Proliferation of A549 cells was influenced by RNAi
of LASP-1, and the effect was time-dependent; with the
increase in treatment time, the negative regulatory effect of
LASP-1-siRNA became evident (Fig.
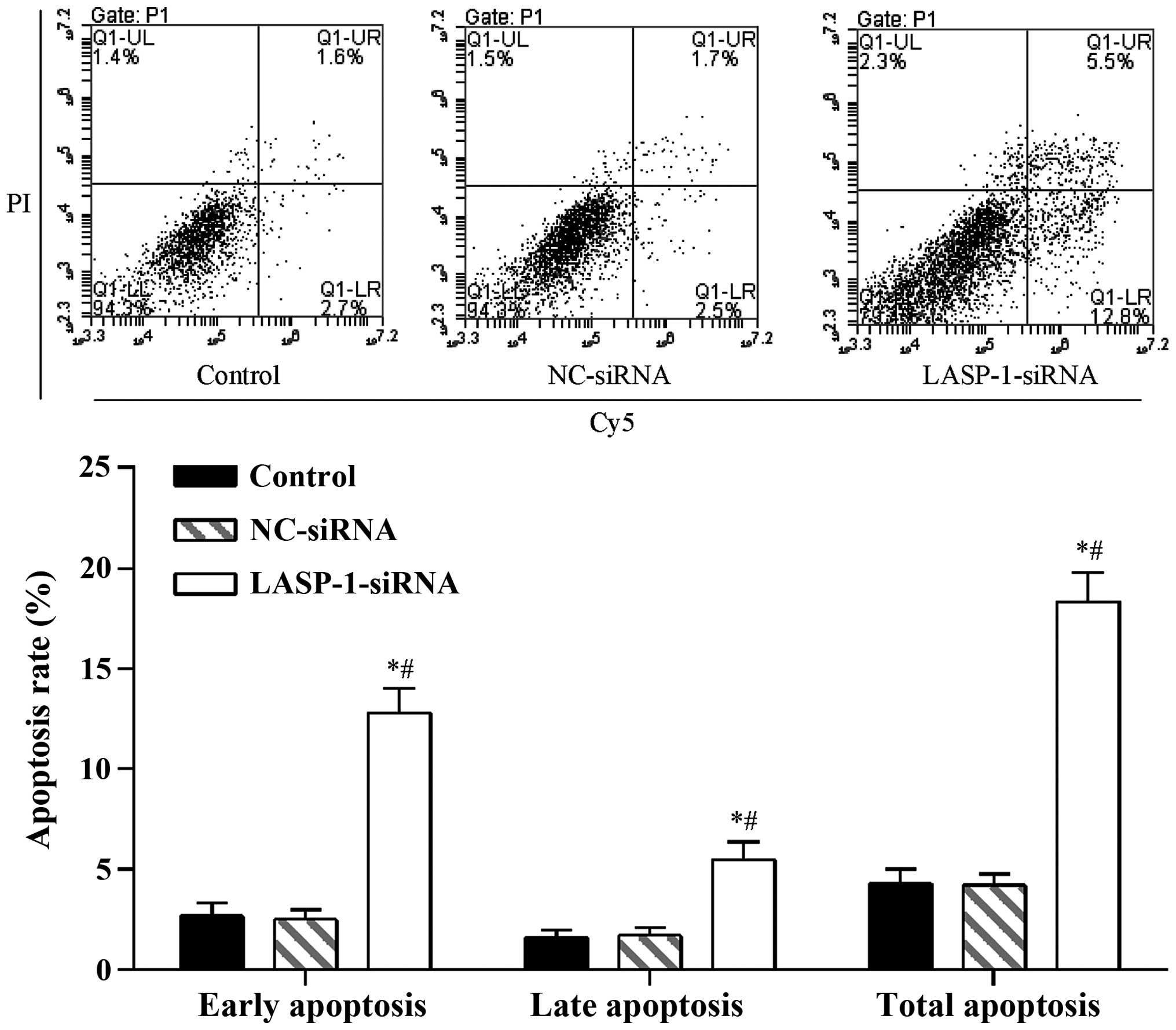
3). A similar pattern was also observed in the FACS results,
where RNAi with LASP-1-specific siRNA significantly
increased the apoptosis percentage (18.3%) of the A549 cell line
compared with the other two groups (4.3% for the control group and
4.2% for the NC-siRNA group) (Fig.
4). The results of CKK-8 assay and FACS were indicative of the
induction of the autophagy process in lung cancer cells by
downregulation of LASP-1.
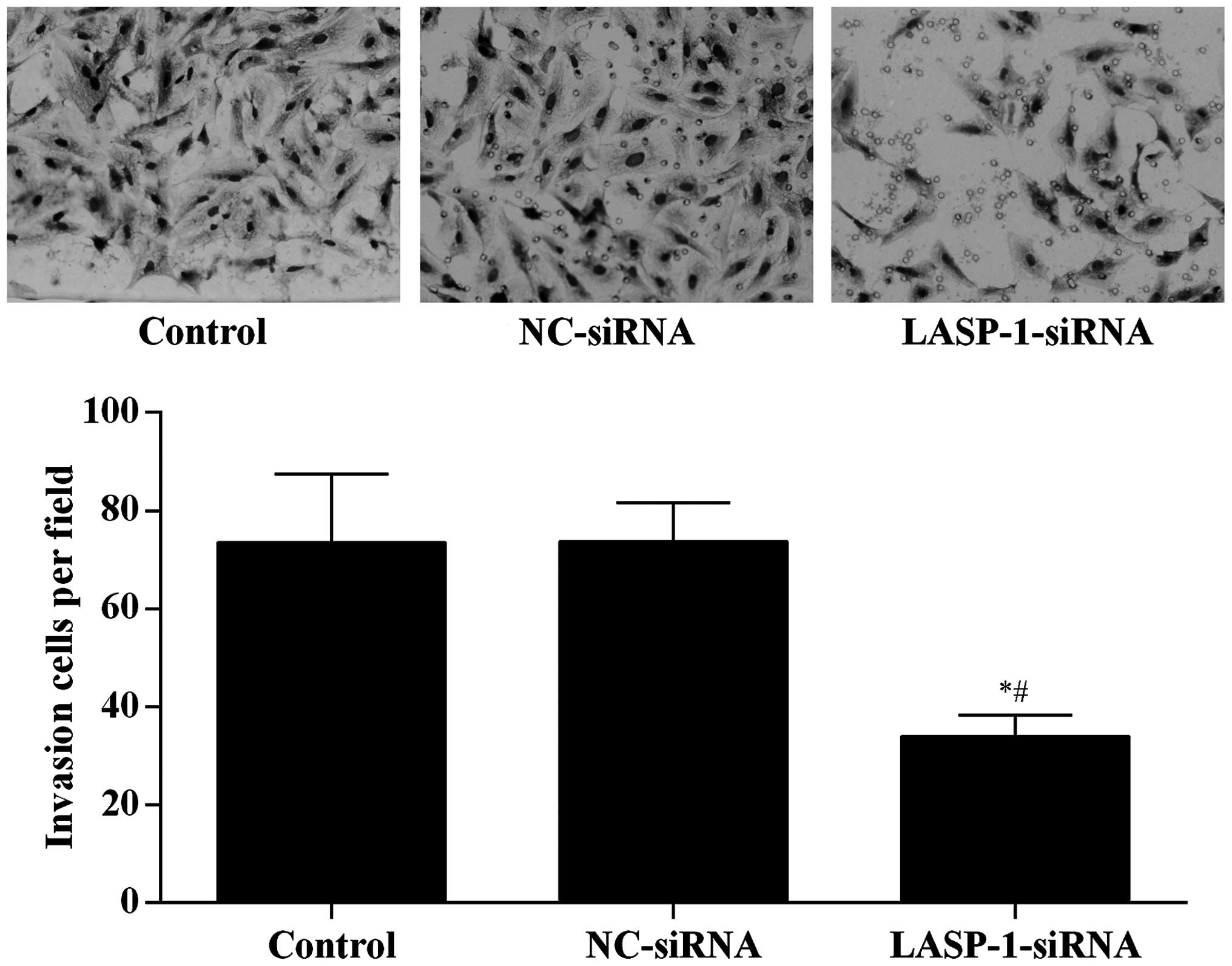
To directly examine the function of LASP-1 on the
migration of A549 cells, Transwell assay was performed in a
modified Boyden chamber with A549 cells transfected with different
treatments. After 4 h, the number of A549 cells that had migrated
through the porous membrane was recorded. Silencing of LASP-1 in
A549 cells significantly reduced cell migration (Fig. 5), suggesting that LASP-1 was necessary
for the metastasis of lung cancer cells.
Discussion
Advances in our understanding of molecular genetics
in lung cancer have led to the identification of key genetic
aberrations, which have potential as therapeutic targets in the
clinic. Currently, molecular subsets identified in lung cancer
include mutation EGFR, KRAS, HER2,
PIK3CA, BRAF and MET genes, and gene
rearrangement in ALK, ROS1 and RET (11). Based on the identification of these
driver oncogenes, lung cancer has been taken as a heterogeneous
disease composed of various molecular subsets. In the present
study, we investigated the expression of another tumor-associated
factor, LASP-1, in lung cancer tissues for the first time. We
observed that LASP-1 mRNA and protein levels were significantly
increased in lung cancer tissues compared with para-carcinoma
tissues. By silencing the expression of the LASP-1 gene in
the human lung cancer A549 cell line, we established that the
proliferation and migration ability of the cancer cells were
notably weakened while the apoptotic process was induced,
suggesting a key role of LASP-1 in the growth and metastasis of
lung cancer. Our results were consistent with those previous
studies, which revealed the significance of LASP-1 in the
progression and metastasis of a variety of tumor types, including
breast, colorectal, ovarian and hepatocellular cancer (14–18).
To test whether the decreased proliferation of the
A549 cell line could be due to apoptosis, we conducted FACS
analysis. Our data revealed significant cell death in response to
LASP-1-specific siRNA treatment. However, our result was
contrary to the conclusion of Grunewald et al (14), which demonstrated no notable influence
of silencing of LASP-1 on the apoptotic process in BT-20 and MCF-7
cell lines. Although the two studies focused on different tumor
types, the results were indicative of the complex mechanism of
LASP-1 in regulating cancer cell apoptosis.
Cell migration requires temporal and spatial
organization of signal transduction processes that regulate actin
polymerization and focal adhesion turnover. To date, more than 50
different adhesion proteins regulating the rate and organization of
actin polymerization and focal adhesion turnover have been
identified. As a well-known focal adhesion adaptor protein, LASP-1
is closely associated with the cytoskeleton (12,19–21) and
involved in cell migration and invasion. The molecule is capable of
interacting and co-localizing with a series of focal adhesion
proteins, including F-actin, zyxin and LPP. Since cells depleted of
LASP-1 still attach to the extracellular matrix and form focal
adhesions, it is hypothesized that LASP-1 plays a supportive role
in focal adhesion dynamics rather than actual formation of the
related structures (22). Thus,
silencing of LASP-1 may cause a cellular localization change of its
binding partners at the focal adhesion site and influence the
migration ability of cells.
LSAP-1 has also been identified as a p53 target in a
global screen of p53 binding sites (23). The mutation of p53 causes a
loss of tumor suppression function, promoting cellular
proliferation. These mutations in p53 tumor suppressor genes are
reported in over 60% of lung cancer cases (24). A previous study clearly demonstrated
that LASP-1 might be the link for p53 to mediate its function on
the cytoskeleton and in turn alter cell behavior (25). As a result, the p53-LASP-1-focal
adhesion plaque pathway may represent a potential mechanism by
which LASP-1 modulates cell motility and growth. However, the
expression of p53 was not investigated in the current study and so
we are unable to explain the underlying interaction of p53 and
LSAP-1 in lung cancer.
In summary, we quantified the expression of mRNA and
protein of LASP-1 in lung cancer tissues for the first time and
observed an abnormally high level of LASP-1 in lung cancer cells.
After silencing the expression of LASP-1 in the human lung cancer
cell line A549, we noted that the proliferation and migration
ability of the cells were significantly decreased. Moreover, data
from FACS analysis revealed the induction of apoptosis by silencing
LASP-1 in the A549 cell line. Our results indicated the significant
role of LASP-1 in regulating the growth and metastasis of lung
cancer cells. Although we do not have a detailed explanation of the
influence of LASP-1 on lung cancer, the present study inferred that
the molecule had the potential to form a therapy for lung cancer in
the clinic. To elaborate the role of LASP-1 in lung cancer, more
comprehensive studies are due to be conducted in the future.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singh CR and Kathiresan K: Molecular
understanding of lung cancers - a review. Asian Pac J Trop Biomed.
4:(Suppl 1). S35–S41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Novaes FT, Cataneo DC, Junior RL Ruiz,
Defaveri J, Michelin OC and Cataneo AJM: Lung cancer: histology,
staging, treatment and survival. J Bras Pneumol. 34:595–600.
2008.(In English and Portuguese). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Youlden DR, Cramb SM and Baade PD: The
international epidemiology of lung cancer: geographical
distribution and secular trends. J Thorac Oncol. 3:819–831. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson JL, Pillai S and Chellappan SP:
Genetic and biochemical alterations in non-small cell lung cancer.
Biochem Res Int. 2012:9404052014.
|
|
6
|
Rodenhuis S, Slebos RJ, Boot AJ, Evers SG,
Mooi WJ, Wagenaar SS, van Bodegom PC and Bos JL: Incidence and
possible clinical significance of K-ras oncogene activation in
adenocarcinoma of the human lung. Cancer Res. 48:5738–5741.
1988.PubMed/NCBI
|
|
7
|
Liu H, Radisky DC, Yang D, Xu R, Radisky
ES, Bissell MJ and Bishop JM: MYC suppresses cancer metastasis by
direct transcriptional silencing of αv and β3 integrin subunits.
Nat Cell Biol. 14:567–574. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Prins J, De Vries EG and Mulder NH: The
myc family of oncogenes and their presence and importance in
small-cell lung carcinoma and other tumour types. Anticancer Res.
13:1373–1385. 1993.PubMed/NCBI
|
|
9
|
Kurup A and Hanna NH: Treatment of small
cell lung cancer. Crit Rev Oncol Hematol. 52:117–126. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schiller JH, Harrington D, Belani CP,
Langer C, Sandler A, Krook J, Zhu J and Johnson DH: Eastern
Cooperative Oncology Group: Comparison of four chemotherapy
regimens for advanced non-small-cell lung cancer. N Engl J Med.
346:92–98. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumarakulasinghe NB, van Zanwijk N and Soo
RA: Molecular targeted therapy in the treatment of advanced stage
non-small cell lung cancer (NSCLC). Respirology. 20:370–378. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chew CS, Chen X, Parente JA, Tarrer S,
Okamoto C and Qin HY: Lasp-1 binds to non-muscle F-actin in vitro
and is localized within multiple sites of dynamic actin assembly in
vivo. J Cell Sci. 115:4787–4799. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nakagawa H, Terasaki AG, Suzuki H, Ohashi
K and Miyamoto S: Short-term retention of actin filament binding
proteins on lamellipodial actin bundles. FEBS Lett. 580:3223–3228.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grunewald TG, Kammerer U, Schulze E,
Schindler D, Honig A, Zimmer M and Butt E: Silencing of LASP-1
influences zyxin localization, inhibits proliferation and reduces
migration in breast cancer cells. Exp Cell Res. 312:974–982. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao L, Wang H, Liu C, Liu Y, Wang X, Wang
S, Sun X, Li J, Deng Y, Jiang Y and Ding Y: Promotion of colorectal
cancer growth and metastasis by the LIM and SH3 domain protein 1.
Gut. 59:1226–1235. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Salvi A, Bongarzone I, Miccichè F, Arici
B, Barlati S and De Petro G: Proteomic identification of LASP-1
down-regulation after RNAi urokinase silencing in human
hepatocellular carcinoma cells. Neoplasia. 11:207–219. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Li W, Jin X, Cui S and Zhao L: LIM
and SH3 protein 1, a promoter of cell proliferation and migration,
is a novel independent prognostic indicator in hepatocellular
carcinoma. Eur J Cancer. 49:974–983. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiyomaru T, Enokida H, Kawakami K,
Tatarano S, Uchida Y, Kawahara K, Nishiyama K, Seki N and Nakagawa
M: Functional role of LASP1 in cell viability and its regulation by
microRNAs in bladder cancer. Urol Oncol. 30:434–443. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Butt E, Gambaryan S, Göttfert N, Galler A,
Marcus K and Meyer HE: Actin binding of human LIM and SH3 protein
is regulated by cGMP-and cAMP-dependent protein kinase
phosphorylation on serine 146. J Biol Chem. 278:15601–15607. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li B, Zhuang L and Trueb B: Zyxin
interacts with the SH3 domains of the cytoskeletal proteins
LIM-nebulette and Lasp-1. J Biol Chem. 279:20401–20410. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schreiber V, Moog-Lutz C, Régnier CH,
Chenard MP, Boeuf H, Vonesch JL, Tomasetto C and Rio MC: Lasp-1, a
novel type of actin-binding protein accumulating in cell membrane
extensions. Mol Med. 4:675–687. 1998.PubMed/NCBI
|
|
22
|
Chew CS, Parente JA Jr, Zhou CJ, Baranco E
and Chen X: Lasp-1 is a regulated phosphoprotein within the cAMP
signaling pathway in the gastric parietal cell. Am J Physiol.
275:C56–C67. 1998.PubMed/NCBI
|
|
23
|
Wei CL, Wu Q, Vega VB, Chiu KP, Ng P,
Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al: A global map of
p53 transcription-factor binding sites in the human genome. Cell.
124:207–219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pfeifer GP, Denissenko MF, Olivier M,
Tretyakova N, Hecht SS and Hainaut P: Tobacco smoke carcinogens,
DNA damage and p53 mutations in smoking-associated cancers.
Oncogene. 21:7435–7451. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang B, Feng P, Xiao Z and Ren EC: LIM and
SH3 protein 1 (Lasp1) is a novel p53 transcriptional target
involved in hepatocellular carcinoma. J Hepatol. 50:528–537. 2009.
View Article : Google Scholar : PubMed/NCBI
|