Introduction
Esophageal carcinoma is the fifth and eighth most
common cause of mortality worldwide in men and women, respectively
(1). In Eastern Asia, esophageal
squamous cell carcinoma (ESCC) is the predominant type of
esophageal carcinoma, with >100 cases/100,000 population
reported annually (2). ESCC is also
the major histological type in Chinese populations, resulting in
150,000 mortalities annually (3).
Despite the availability of multiple treatments for ESCC, surgical
resection remains the primary choice for non-metastatic cases
(4). However, even upon curative
surgery, the 5-year survival rate for ESCC patients is only
26.2–49.4%, due to local or distant recurrences (5). Therefore, it is critical to identify
novel molecular mechanisms to elucidate ESCC oncogenesis and
metastasis.
MicroRNAs (miRs) are small endogenous non-coding
RNAs that play vital roles in various biological processes
(6,7).
Mature microRNAs usually bind to 3′-untranslated regions (3′-UTRs)
of target genes, leading to messenger RNA degradation or repression
of translation, thus downregulating the expression of target genes
at post-transcriptional levels (6,8). Since the
first microRNA (microRNA-lin-4) was identified in 1993, an
increasing number of microRNAs have been reported to be involved in
numerous physiological and pathological processes, including
carcinogenesis (9,10). Several microRNAs, including miR-21,
miR-34a and miR-155, have been observed to be associated with
carcinogenesis by targeting oncogenes or anti-oncogenes (11–13).
In the present study, the expression of miR-193a-3p
was examined in ESCC specimens, and the function of miR-193a-3p in
ESCC cells was investigated via cell migration, proliferation and
apoptosis assays. The results revealed that miR-193a-3p was
upregulated in ESCC tissues and functioned as an oncogene in ESCC
by affecting cellular migration, proliferation and apoptosis.
Materials and methods
ESCC tissue samples collection
All ESCC tissues and adjacent normal tissues used in
the present study were collected from Dongnan Affiliated Hospital
of Xiamen University (Zhangzhou, China) between February, 2013 and
December, 2014. Written informed consent was obtained from all
patients. The collection and use of these samples was approved by
the Ethics Committee of Dongnan Affiliated Hospital of Xiamen
University. Fresh ESCC and adjacent normal tissues were immersed in
RNAlater (Qiagen GmbH, Hilden, Germany) in 30 min following
resection, and subsequently stored at −80°C.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
According to the manufacturer's protocol, total RNA
was extracted with TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and purified with RNeasy Maxi
kit (Qiagen GmbH). Total RNA (1 µg of each sample) was used for RT
using miScript RT kit (Qiagen GmbH) to obtain the complementary DNA
(cDNA) templates. qPCR reaction of miR-193a-3p was performed in an
ABI PRISM® 7000 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using miScript SYBR
Green PCR kit (Qiagen GmbH). U6 was used as an endogenous control.
The 20 µl reaction mixture contained 2X QuantiTect SYBR Green PCR
Master mix (10 µl), 10X miScript Universal Primer (2 µl), specific
microRNA primer (0.4 µl), cDNA template (1 µl) and RNase-free
water. The forward primer of miR-193a-3p was
5′-AACTGGCCTACAAAGTCCCAGT-3′ and the reverse primer was provided in
the miScript SYBR Green PCR kit. The forward primer and reverse
primers of U6 were 5′-CTCGCTTCGGCAGCACA-3′ and
5′-ACGCTTCACGAATTTGCGT-3′, respectively. Amplification conditions
were set as: 95°C for 2 min, followed by 40 cycles of 94°C for 15
sec, 58°C for 30 sec and 72°C for 30 sec.
Cell culture and transfection
The human ESCC cell lines Eca109 and TE-1 used in
the present study were purchased from the Shanghai Institute of
Biochemistry and Cell Biology (Shanghai, China), and were cultured
in RPMI 1640 (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 g/ml streptomycin at
37°C in a humidified incubator containing 5% CO2. For
the downregulation of miR-193a-3p expression, a synthesized
miR-193a-3p inhibitor (Shanghai GenePharma Co., Ltd., Shanghai,
China) was transfected into the cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The transfection efficiency and changes in
miR-193a-3p expression were determined by fluorescence microscopy
and RT-qPCR, respectively.
Migration assay
The migratory ability of ESCC cells (Eca109 and
TE-1) in vitro was assessed by wound scratch assay.
Approximately 150,000 cells were seeded in a 12-well dish and were
transfected with miR-193a-3p inhibitor (60 pmol) or negative
control (60 pmol) with Lipofectamine 2000 24 h later. After 5 h of
transfection, a vertical wound was created with the tip of a
sterile 10-µl pipette, and markers were assigned to enable the
observation of cells in the correct location. The cells were then
rinsed three times with phosphate-buffered saline (PBS) and
incubated at 37°C. Images of the wound scratches were captured with
a digital camera system at 0 and 24 h after the wounds were made at
the same location. Wound widths (µm) were measured with a standard
caliper, and the experiments were performed in triplicate and
analyzed by at least two observers.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell proliferation of ESCC lines (Eca109 and TE-1)
was assessed with an MTT assay kit (Sigma-Aldrich, St. Louis, MO,
USA). Approximately 5,000 cells were seeded into 96-well plates and
transfected with 5 pmol of miR-193a-3p inhibitor or negative
control. Next, 20 µl of MTT (5 mg/ml; Sigma-Aldrich) was added to
the culture medium of each well at 0, 24, 48 and 72 h
post-transfection. After incubation for 4 h, the MTT medium was
removed, and 150 µl of dimethyl sulfoxide was added. After shaking
for 15 min at room temperature, the optical density (OD) of each
sample was assessed with an enzyme-linked immunosorbent assay
instrument (model 680; Bio-Rad Rad Laboratories, Inc., Hercules,
CA, USA) at a wavelength of 490/630 nm.
Flow cytometry
For apoptosis assay, ESCC cells (Eca109 and TE-1)
were cultured in 6-well plates at 37°C and transfected with
miR-193a-3p inhibitor or negative control at a confluence of ~65%.
After 48 h of transfection, cells were harvested and washed twice
with cold PBS, resuspended in 10 µl 1X binding buffer (Invitrogen;
Thermo Fisher Scientific, Inc.). Subsequently, 5 µl of Annexin
V-fluorescein isothiocyanate (Invitrogen; Thermo Fisher Scientific,
Inc.) and 10 µl of propidium iodide were added to each sample. The
fluorescence of the stained cells was then analyzed by flow
cytometry (Beckman Coulter, Inc., Brea, CA, USA) using an
excitation wavelength of 488 nm within 30 min of staining,
according to the manufacturer's protocol.
Statistical analysis
Statistical analysis was conducted with SPSS 17.0
statistical software package (SPSS, Inc., Chicago, IL, USA).
Statistical significance was determined with paired t-test
and Student's t test. P<0.05 was considered to indicate a
statistically significant difference.
Results
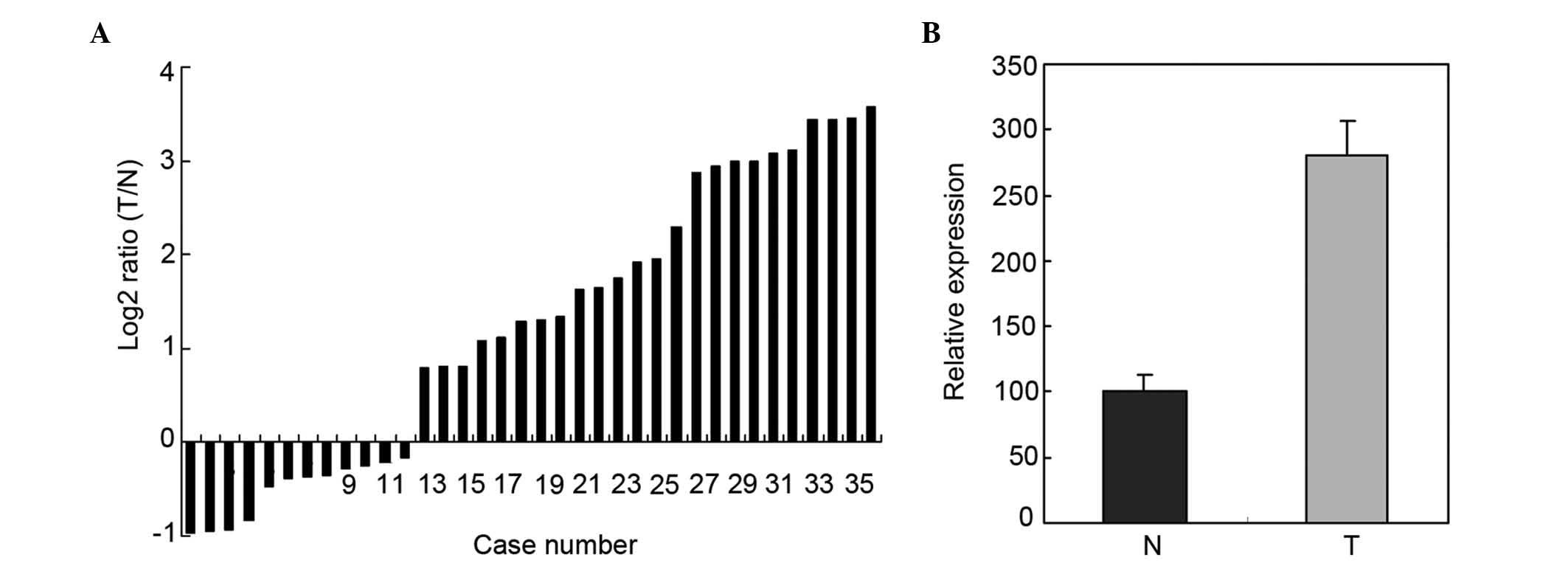
Upregulation of miR-193a-3p in 36
paired ESCC tissues and adjacent normal tissues by RT-qPCR
RT-qPCR was used to determine the expression of
miR-193a-3p in 36 paired ESCC tissues and adjacent normal tissues.
The relative expression of miR-193a-3p is represented in Fig. 1A. As shown in Fig. 1B, the miR-193a-3p expression in ESCC
tissues was significantly higher than that in adjacent normal
tissues (P=0.0153).
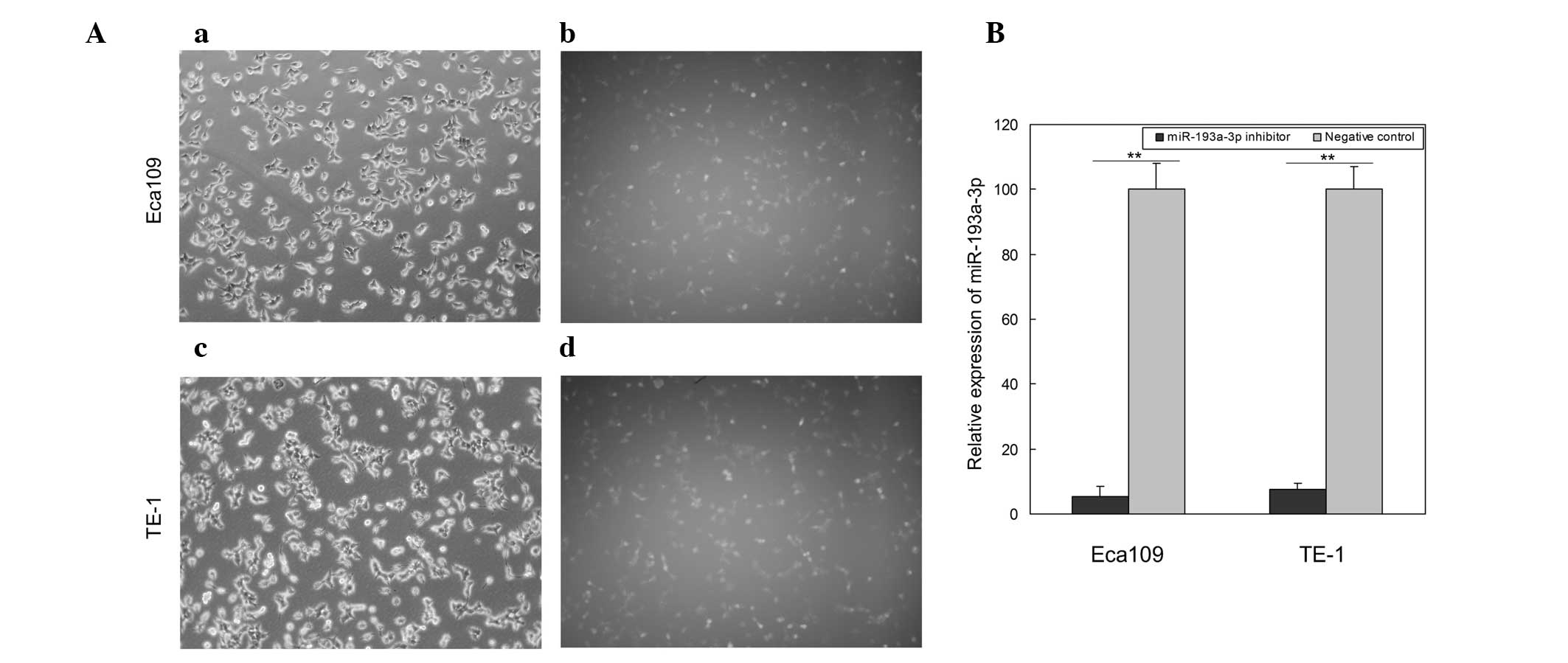
Transfection and inhibition
efficiency
To determine the function of miR-193a-3p in ESCC,
miR-193a-3p inhibitor and negative control were transfected into
the ESCC cell lines Eca109 and TE-1. Images of the cells
transfected with fluorescein amidite-labeled negative control were
obtained at 6 h post-transfection, and revealed that the
transfection efficiency was ~85 and 80% in Eca109 and TE-1 cells,
respectively (Fig. 2A). Compared with
the negative control, the relative expression of miR-193a-3p in
Eca109 and TE-1 cells transfected with miR-193a-3p inhibitor was
5.5 and 7.3%, respectively (Fig. 2B).
These results suggested that the miR-193a-3p inhibitor used in the
present study was able to effectively downregulate the expression
of miR-193a-3p.
Downregulation of miR-193a-3p
suppresses ESCC cell migration in vitro
Wound scratch assay was performed to determine the
effects of miR-193a-3p on ESCC cell migration in vitro.
Compared with the negative control group, the wound widths of
Eca109 and TE-1 cells transfected with miR-193a-3p inhibitor were
significantly wider (P=0.0322 and 0.0306, respectively; Fig. 3), which indicated that downregulation
of miR-193a-3p inhibited the migration of ESCC cells (Fig. 3).
Reduction of miR-193a-3p inhibits cell
proliferation
The impact of miR-193a-3p on cell proliferation in
ESCC was analyzed by MTT assay. The OD values of miR-193a-3p
inhibitor group and negative control group were measured at 0, 24,
48 and 72 h post-transfection. The results indicated that the
proliferation of Eca109 cells decreased by 6.45% (P=0.0318), 10.63%
(P=0.0131) and 14.19% (P=0.0019), while the proliferation of TE-1
cells decreased by 6.78% (P=0.0356), 11.51% (P=0.0152) and 14.72%
(P=0.0025), at 24, 48 and 72 h post-transfection, respectively
(Fig. 4). This indicated that
downregulation of miR-193a-3p suppressed the proliferation of ESCC
cells in vitro.
Inhibition of miR-193a-3p promotes
ESCC cell apoptosis
To determine the function of miR-193a-3p on ESCC
cell apoptosis, flow cytometry was performed to detect the
apoptosis rates upon transfection. As represented in Fig. 5, the apoptosis rates of Eca109 cells
transfected with miR-193a-3p inhibitor and negative control were
7.2 and 2.7% (P=0.0121), respectively, following transfection for
48 h. The apoptosis rates of TE-1 cells were 6.6 and 2.8%
(P=0.0154) upon transfection with miR-193a-3p inhibitor and
negative control, respectively. These results demonstrated that
inhibition of miR-193a-3p induced ESCC cell apoptosis.
Discussion
Carcinogenesis involves the activation of a number
of oncogenes and the inactivation of a number of anti-oncogenes
(14). In the complicated regulatory
network of oncogenes and anti-oncogenes, microRNAs plays a vital
role by controlling the expression of target genes at
post-transcriptional levels (15). By
regulating the levels of oncogenes or anti-oncogenes, a microRNA
can act as an anti-oncogene or as an oncogene (8). Although only accounting for a small
fraction of the expressed genome, microRNAs play crucial roles in
diverse cellular processes, including cell proliferation, cellular
differentiation, cell death, metabolism, apoptosis, motility,
invasion and morphogenesis (6,10,16–20). A
number of microRNAs were observed to be upregulated in ESCC,
including miR-21 (21–23), miR-205 (24), miR-10b (25), miR-23a (26), miR-26a (26), miR-27b (26), miR-96 (26), miR-128b (26), miR-129 (26), miR-93 (23), miR-192 (23) and miR-194 (23). In addition, downregulation of miR-375
(21), miR-203 (23), miR-205 (23), miR-27b (23), miR-125b (23) and miR-100 (23) expression has been detected in ESCC.
Furthermore, numerous microRNAs were noticed to play the role of
oncogene or anti-oncogene in ESCC, including miR-21, which
facilitates ESCC growth by targeting phosphatase and tensin homolog
and programmed cell death 4 (22,27);
miR-296, which contributes to ESCC growth by targeting cyclin D1
and p27 (28); miR-210, which targets
fibroblast growth factor receptor-like 1, thus exerting a negative
effect on cell cycle and proliferation (29); miR-145, miR-133a and miR-133b, which
converge to target fascin 1, thus reducing cell growth and invasion
(30); and miR-593, which may
contribute to carcinogenesis through polo-like kinase 1 (31).
miR-193a-3p can function as a tumor suppressor,
since it inhibits cell-cycle progression and proliferation in
breast cancer through targeting epidermal growth factor
receptor-driven cell-cycle network proteins (32) and induces apoptosis in both U-251 and
HeLa cells (33). In addition,
miR-193a-3p suppresses the metastasis of human non-small-cell lung
cancer (34,35). However, the expression and role of
miR-193a-3p in ESCC remains unclear.
To determine the expression and role of miR-193a-3p
in ESCC, RT-qPCR was used in the present study to quantify the
miR-193a-3p level in 36 cases of ESCC tissues and paired normal
tissues. The present study demonstrated that miR-193a-3p expression
was significantly upregulated in ESCC tissues, compared with its
expression levels in paired normal esophageal tissues. The effects
of miR-193a-3p on ESCC cell migration, proliferation and apoptosis
were then analyzed by transfection of ESCC cell lines with a
synthetic miR-193a-3p inhibitor. Transfection of miR-193a-3p
inhibitor into the ESCC cell lines Eca109 and TE-1 inhibited
cellular proliferation and migration and induced apoptosis,
compared with the negative control group. These findings suggested
that miR-193a-3p may act as a oncogene in ESCC by inhibiting
cellular proliferation and migration and by promoting cellular
apoptosis. Further studies on miR-193a-3p target genes are required
to clarify the mechanism of action of miR-193a-3p in ESCC.
The role of miR-193a-3p in different cancers appears
to be controversial, as it was identified as a tumor suppressor in
certain cancers (32–35) and as an oncogene in ESCC, according to
the results of the present study. Regardless of experimental error,
this contradiction may be explained as ‘imperfect’ complementary
interactions between microRNAs and their target genes. Contrary to
small interfering RNA, the interactions between microRNAs and the
3′-UTRs of their target genes are not always perfectly
complementary (particularly in mammals), which leads to relative
rather than complete specificity between microRNAs and their target
genes (36). Accurate interactions
between microRNAs and their target genes may be further dictated by
cell types and the microenvironment, thus contributing to the
divergent regulatory roles of certain microRNAs (6,37).
In conclusion, the present study revealed that
miR-193a-3p was upregulated in ESCC and played a vital oncogenic
role in ESCC by affecting cellular migration, proliferation and
apoptosis. Further studies are required to define its mechanism of
action in ESCC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao P, Dai M, Chen W and Li N: Cancer
trends in China. Jpn J Clin Oncol. 40:281–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han LH, Jia YB, Song QX, Wang JB, Wang NN
and Cheng YF: Prognostic significance of preoperative
lymphocyte-monocyte ratio in patients with resectable esophageal
squamous cell carcinoma. Asian Pac J Cancer Prev. 16:2245–2250.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu J, Xie X, Zhou C, Peng S, Rao D and Fu
J: Which factors are associated with actual 5-year survival of
oesophageal squamous cell carcinoma? Eur J Cardiothorac Surg.
41:e7–e11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang L, Liu X, Chen Z, Jin Y, Heidbreder
CE, Kolokythas A, Wang A, Dai Y and Zhou X: MicroRNA-7 targets
IGF1R (insulin-like growth factor 1 receptor) in tongue squamous
cell carcinoma cells. Biochem J. 432:199–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Soeda S, Ohyashiki JH, Ohtsuki K, Umezu T,
Setoguchi Y and Ohyashiki K: Clinical relevance of plasma miR-106b
levels in patients with chronic obstructive pulmonary disease. Int
J Mol Med. 31:533–539. 2013.PubMed/NCBI
|
|
8
|
Xiong Y, Zhang L, Holloway AK, Wu X, Su L
and Kebebew E: MiR-886-3p regulates cell proliferation and
migration and is dysregulated in familial non-medullary thyroid
cancer. PLoS One. 6:e247172011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ha TY: MicroRNAs in Human Diseases: From
cancer to Cardiovascular Disease. Immune Netw. 11:135–154. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhai Q, Zhou L, Zhao C, Wan J, Yu Z, Guo
X, Qin J, Chen J and Lu R: Identification of miR-508-3p and
miR-509-3p that are associated with cell invasion and migration and
involved in the apoptosis of renal cell carcinoma. Biochem Biophys
Res Commun. 419:621–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shibuya H, Iinuma H, Shimada R, Horiuchi A
and Watanabe T: Clinicopathological and prognostic value of
microRNA-21 and microRNA-155 in colorectal cancer. Oncology.
79:313–320. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akao Y, Noguchi S, Iio A, Kojima K, Takagi
T and Naoe T: Dysregulation of microRNA-34a expression causes
drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer
Lett. 300:197–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tili E, Michaille JJ, Wernicke D, Alder H,
Costinean S, Volinia S and Croce CM: Mutator activity induced by
microRNA-155 (miR-155) links inflammation and cancer. Proc Natl
Acad Sci USA. 108:4908–4913. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y,
Yu W, Wu X, Ye J, Yang S, et al: Identification of miR-7 as an
oncogene in renal cell carcinoma. J Mol Histol. 44:669–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jing Q, Huang S, Guth S, Zarubin T,
Motoyama A, Chen J, Di Padova F, Lin SC, Gram H and Han J:
Involvement of microRNA in AU-rich element-mediated mRNA
instability. Cell. 120:623–634. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bhattacharyya S, Balakathiresan NS,
Dalgard C, Gutti U, Armistead D, Jozwik C, Srivastava M, Pollard HB
and Biswas R: Elevated miR-155 promotes inflammation in cystic
fibrosis by driving hyperexpression of interleukin-8. J Biol Chem.
286:11604–11615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lucotti S, Rainaldi G, Evangelista M and
Rizzo M: Fludarabine treatment favors the retention of miR-485-3p
by prostate cancer cells: Implications for survival. Mol Cancer.
12:522013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sarver AL, French AJ, Borralho PM,
Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM,
Boardman LA, Cunningham JM, et al: Human colon cancer profiles show
differential microRNA expression depending on mismatch repair
status and are characteristic of undifferentiated proliferative
states. BMC Cancer. 9:4012009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H
and Zhou X: MicroRNA-222 regulates cell invasion by targeting
matrix metalloproteinase 1 (MMP1) and manganese superoxide
dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines.
Cancer Genomics Proteomics. 6:131–139. 2009.PubMed/NCBI
|
|
21
|
Mathe EA, Nguyen GH, Bowman ED, Zhao Y,
Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et
al: MicroRNA expression in squamous cell carcinoma and
adenocarcinoma of the esophagus: Associations with survival. Clin
Cancer Res. 15:6192–6200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M
and Baba H: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260; discussion 260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Matsushima K, Isomoto H, Kohno S and Nakao
K: MicroRNAs and esophageal squamous cell carcinoma. Digestion.
82:138–144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H
and Liu Z: MicroRNA-10b promotes migration and invasion through
KLF4 in human esophageal cancer cell lines. J Biol Chem.
285:7986–7994. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ogawa R, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Katada T, Harata K, Tanaka T and Fujii Y: Expression
profiling of micro-RNAs in human esophageal squamous cell carcinoma
using RT-PCR. Med Mol Morphol. 42:102–109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma WJ, Lv GD, Tuersun A, Liu Q, Liu H,
Zheng ST, Huang CG, Feng JG, Wang X, Lin RY, et al: Role of
microRNA-21 and effect on PTEN in Kazakh's esophageal squamous cell
carcinoma. Mol Biol Rep. 38:3253–3260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hong L, Han Y, Zhang H, Li M, Gong T, Sun
L, Wu K, Zhao Q and Fan D: The prognostic and chemotherapeutic
value of miR-296 in esophageal squamous cell carcinoma. Ann Surg.
251:1056–1063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsuchiya S, Fujiwara T, Sato F, Shimada Y,
Tanaka E, Sakai Y, Shimizu K and Tsujimoto G: MicroRNA-210
regulates cancer cell proliferation through targeting fibroblast
growth factor receptor-like 1 (FGFRL1). J Biol Chem. 286:420–428.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M and
Matsubara H: miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ito T, Sato F, Kan T, Cheng Y, David S,
Agarwal R, Paun BC, Jin Z, Olaru AV, Hamilton JP, et al: Polo-like
kinase 1 regulates cell proliferation and is targeted by miR-593*
in esophageal cancer. Int J Cancer. 129:2134–2146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Uhlmann S, Mannsperger H, Zhang JD, Horvat
EÁ, Schmidt C, Küblbeck M, Henjes F, Ward A, Tschulena U, Zweig K,
et al: Global microRNA level regulation of EGFR-driven cell-cycle
protein network in breast cancer. Mol Syst Biol. 8:5702012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kwon JE, Kim BY, Kwak SY, Bae IH and Han
YH: Ionizing radiation-inducible microRNA miR-193a-3p induces
apoptosis by directly targeting Mcl-1. Apoptosis. 18:896–909. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deng W, Yan M, Yu T, Ge H, Lin H, Li J,
Liu Y, Geng Q, Zhu M, Liu L, et al: Quantitative Proteomic analysis
of the metastasis-inhibitory mechanism of miR-193a-3p in non-small
cell lung cancer. Cell Physiol Biochem. 35:1677–1688. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kharaziha P, Ceder S, Li Q and Panaretakis
T: Tumor cell-derived exosomes: A message in a bottle. Biochim
Biophys Acta. 1826:103–111. 2012.PubMed/NCBI
|