Introduction
The high incidence of gastric cancer (GC) and its
associated mortality pose severe threats to human health (1,2). Although
curative gastrectomy followed by adjuvant therapy has been
demonstrated to prolong the survival of patients with stage II/III
GC, certain patients develop locoregional or distant recurrence
(3–5).
Patients with stage IV GC almost always possess a poor prognosis
(6,7).
Identifying biomarkers relevant to the recurrence and metastasis of
GC may assist clinicians in tailoring therapies by identifying
high-risk patients and proposing novel molecular targets for the
treatment of GC.
Recent analysis of gene and protein expression
profiles, as well as oncogenic signaling pathways, suggests the
existence of molecular subtypes of GC (8–10). This
molecular diversity leads to clinical heterogeneity (8). Although GCs represent a biologically
heterogeneous group of diseases, treatment strategy is generally
determined by clinical stage alone, with no consideration of the
molecular characteristics of the cancer (2). Detailed molecular characterization of a
patient's tumor may enable tailored therapies that improve the
likelihood of a positive outcome and decrease toxicity.
SAM domain, SH3 domain and nuclear localization
signals 1 (SAMSN1) encodes one of a family of SH3-domain
containing cytoplasmic adaptor proteins expressed in lymphocytes
(11,12). SAMSN1 is mainly expressed by
hematopoietic cells and mediates B-cell activation and
differentiation. The SAMSN1 gene is located on chromosome
21q11-21, within a region associated with heterozygous deletions
that is frequently present in lung cancer cells, suggesting that
SAMSN1 acts as a tumor suppressor (13,14). This
possibility is supported by the study of Noll et al
(15), which revealed that
SAMSN1 is a suppressor of multiple myeloma (15). To date, the precise role of
SAMSN1 in oncogenesis remains to be fully elucidated,
particularly in cancer of the digestive tract, including GC. The
present study hypothesized that the dysregulation or absence of
SAMSN1 expression contributes to the initiation and
progression of GC. The aims of the present study were to
investigate the clinical significance of SAMSN1 expression,
define the mechanism of SAMSN1 transcriptional regulation,
establish whether SAMSN1 contributes to tumorigenesis and
assess the clinical utility of SAMSN1 as a potential
prognostic marker and as a target for therapy in GC.
Materials and methods
Cell lines and tissue samples
The GC cell lines MKN1, MKN45, MKN74, NUGC2, NUGC3,
NUGC4 and SC-6-JCK were obtained from the Japanese Collection of
Research Bioresources Cell Bank (Osaka, Japan). The AGS, KATOIII
and N87 cell lines were acquired from the American Type Culture
Collection (Manassas, VA, USA). The GCIY was obtained from Tohoku
University, Sendai, Japan. A control, non-tumorigenic epithelial
cell line (FHs 74) was purchased from the American Type Culture
Collection. Cells were cultured in RPMI-1640 medium (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Thermo Fisher Scientific, Inc.) and maintained in a
5% CO2 atmosphere at 37°C. For FHs 74 cells, the medium
was additionally supplemented with 30 ng/ml epidermal growth factor
(Sigma-Aldrich; EMD Millipore, Billerica, MA, USA). Total RNA was
extracted using an RNeasy Mini kit (Qiagen GmbH, Hilden, Germany)
and used as a template for the generation of complementary DNA as
described previously (16,17). Primary GC tissues and corresponding
normal adjacent tissues were collected from 175 patients who
underwent gastric resection for GC without neoadjuvant therapy at
Nagoya University Hospital (Nagoya, Japan) between November 2001
and December 2012. Patients who received neoadjuvant therapy were
excluded, as it was difficult to obtain cancer cells from scarred
tissues. Following collection, tissue samples were immediately
frozen in liquid nitrogen and stored at −80°C until the time of RNA
extraction. Corresponding normal adjacent gastric mucosa samples
were obtained from each patient and were collected from a region no
less than 5 cm from the tumor edge. To determine whether the
expression status of SAMSN1 differed according to tumor
histology, patients were categorized into two histological
subtypes: Differentiated (papillary, well differentiated and
moderately differentiated adenocarcinoma) and undifferentiated
(poorly differentiated adenocarcinoma, signet ring cell carcinoma
and mucinous carcinoma) (18). Since
2006, adjuvant chemotherapy using S-1 (an oral fluorinated
pyrimidine) has been administered to all Union for International
Cancer Control (UICC) stage II/III GC patients (unless
contraindicated by the patient's condition) (19,20).
Patients were followed-up at least once every 3 months for 2 years
following surgery, and then every 6 months for 5 years or until
death. Physical examination, laboratory tests and enhanced computed
tomography (chest and abdominal cavity) were performed at each
visit (21). The chemotherapy regimen
for patients with distant metastasis or recurrence was chosen at
the physician's discretion. The present study conformed to the
ethical guidelines of the World Medical Association Declaration of
Helsinki: Ethical Principles for Medical Research Involving Human
Subjects, and was approved by the Institutional Review Board of
Nagoya University, Nagoya, Japan. Written informed consent for
usage of clinical samples and data, as required by the
institutional review board, was obtained from all patients
(22).
SAMSN1 mRNA expression analysis
SAMSN1 mRNA expression levels in 11 GC cell
lines and 175 primary GC tissues and corresponding normal adjacent
tissues were analyzed by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) using an ABI StepOnePlus
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) in conjunction with the gene specific primers listed in
Table I. Cycling conditions were as
follows: One cycle at 95°C for 10 min, 40 cycles at 95°C for 5 sec
and 60°C for 60 sec To investigate the oncological role of
SAMSN1 in GC, correlation analysis was performed to evaluate
the association between the pattern of SAMSN1 mRNA
expression and clinicopathological parameters, including patient
survival following gastrectomy. Each of the 175 patients was
assigned to one of two groups (low and high SAMSN1
expression) according to their median level of SAMSN1 mRNA
expression in GC tissues. Additionally, the prognostic impact of
SAMSN1 mRNA expression on patients categorized according to
the 7th UICC staging system was also evaluated (23).
 | Table I.Primers and associated annealing
temperatures. |
Table I.
Primers and associated annealing
temperatures.
| Gene | Experiment | Direction | Sequence,
5′-3′ | Product size,
bp | Annealing
temperature, °C |
|---|
| SAMSN1 | RT-qPCR | Forward |
TGCTCAAGAGAAAGCCATCC | 97 | 60 |
|
|
| Reverse |
TTATTCCGAAAACGATCGAAA |
|
|
|
| Bisulfite | Forward |
TTGTTTTTATTTTGAGTTGTGTTTGT | 416 | 62 |
|
| Sequencing 1 | Reverse |
ACTAAACTTCCTCCATTACTCTCTCTC |
|
|
|
| Bisulfite | Forward |
AGTTATGTTTTTATTTATATTTAGAATGGG | 257 | 64 |
|
| Sequencing 2 | Reverse |
TCACCCAAACTAAAATACAATAACA |
|
|
| GAPDH | RT-qPCR | Forward |
GAAGGTGAAGGTCGGAGTC | 226 | 60 |
|
|
| Probe |
CAAGCTTCCCGTTCTCAGCC |
|
|
|
|
| Reverse |
GAAGATGGTGATGGGATTTC |
|
|
Bisulfite sequence analysis
Genomic DNA from GC cell lines was treated with
bisulfite using the EpiTect Bisulfate kits (Qiagen GmbH) and
sequenced to determine the levels of DNA methylation according to
previously published procedures (24).
Immunohistochemistry
The intensity and pattern of SAMSN1 protein
expression was determined by immunohistochemical staining using 48
representative sections of well-preserved GC tissue as described
previously (25). Sections were
incubated for 1 h at room temperature with a rabbit polyclonal
antibody raised against SAMSN1 (catalog no., 13063-1-AP;
ProteinTech Group, Inc., Chicago, IL, USA) diluted 1:400 in
antibody diluent (Dako, Glostrup, Denmark). The samples were
subsequently washed with phosphate buffered saline, followed by a
10 min incubation with biotinylated rabbit secondary antibody
(Histofine SAB PO(R) kit; Nichirei Corporation, Tokyo, Japan) in a
1:1,000 dilution with ChemMateT antibody diluent (Dako). Sections
were subsequently developed for 3 min using 3,3′- diaminobenzidine
as the substrate (Nichirei Corporation). The patterns of
SAMSN1 staining in GC tissues and corresponding
non-cancerous tissues were compared, and positive blood vessel
staining provided an internal control for the immunolabeling
procedure. Specimens were randomized and coded prior to analysis by
two independent observers blinded to the status of the samples
(26,27).
Statistical analysis
Differences in the relative expression of
SAMSN1 mRNA (normalized to the level of
glyceraldehyde-3-phosphate expression) between the two groups were
analyzed using the Mann-Whitney U test. The χ2 test was
used to analyze the association between the expression status of
SAMSN1 and various clinicopathological parameters. A
correlation between expression patterns of SAMSN1 protein and mRNA
in gastric tissue specimens was also evaluated by the χ2
test. Survival rates were calculated using the Kaplan-Meier method,
and the difference in survival curves was analyzed using the
log-rank test. Multivariate regression analysis was performed to
detect prognostic factors using the Cox proportional hazards model,
and variables with P<0.05 were entered into the final model. All
statistical analysis was performed using JMP version 10 software
(SAS Institute Inc., Cary, NC, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
SAMSN1 expression and methylation
status in GC cell lines
A marked decrease in the level of SAMSN1 mRNA
expression was detected in 8 (73%) of the 11 GC cell lines when
compared with the FHs 74 control cell line. There was no marked
difference in SAMSN1 expression between cell lines derived
from differentiated and undifferentiated GCs (Fig. 1A). No DNA methylation of the
SAMSN1 promoter was detected.
Patient characteristics
The patient population included 134 males and 41
females with an age range from 20–84 years (mean age, 64.7±11.8
years). Pathologically, 106 patients were diagnosed with
undifferentiated GC and 69 with differentiated GC. A total of 39
patients were diagnosed with stage I disease, 29 with stage II, 51
with stage III and 56 with stage IV disease. A total of 119
patients with stage I–III disease underwent R0 resection. A total
of 47/56 patients classified as UICC stage IV were assigned this
diagnosis due to positive peritoneal lavage cytology, localized
peritoneal metastasis or distant lymph node metastasis. A total of
6 of the patients with stage IV disease had synchronous liver
metastasis and a single patient had lung metastasis, and these
individuals underwent gastrectomy to control bleeding or
obstruction to the passage of food.
SAMSN1 mRNA and protein expression in
surgically resected tissues
The mean expression level of SAMSN1 mRNA was
reduced in GC tissues when compared with that in adjacent normal
tissues (P<0.001). However, there was no significant difference
in the expression of SAMSN1 mRNA between patients with
undifferentiated and differentiated GC (Fig. 1B; P=0.067). Immunohistochemical
staining was subsequently performed to investigate the expression
of SAMSN1 protein in those cases where the SAMSN1 mRNA level
in GC tissues was observed to be less or equivalent to that
identified for corresponding non-cancerous tissues. Representative
GC specimens with an increased, equivalent and reduced intensity of
SAMSN1 protein staining in cancerous tissue compared with adjacent
normal tissue are shown in Fig. 2A.
In 48 of the patient samples examined, the pattern of SAMSN1
protein expression correlated significantly with that of the
expression of SAMSN1 mRNA (P=0.005; Fig. 2B).
Prognostic implications of SAMSN1 mRNA
expression
Patients were assigned to one of two groups
according to their median SAMSN1 mRNA expression level in GC
tissues (high expression group, n=87; low expression group, n=88).
Low SAMSN1 mRNA expression was significantly associated with
larger tumor size (>60 mm; P=0.026), but not tumor location or
UICC stage (P=0.639) (Table II).
Patients in the low SAMSN1 expression group were more likely
to have a shorter overall survival time than those in the high
expression group (5-year survival rates were 43% and 66% for the
high and low expression groups, respectively; P=0.004; Fig. 3A). In multivariate analysis for
overall survival, low SAMSN1 mRNA expression was identified
to be an independent prognostic factor (hazard ratio, 1.80; 95%
confidence interval, 1.07–3.05; P=0.025; Table III). When patients were categorized
according to UICC stage, no significant differences in the mean
expression level of SAMSN1 mRNA was observed between groups
(P>0.05, for each), suggesting that SAMSN1 expression was
independent of tumor stage (Fig.
3B).
 | Table II.Association between expression level
of SAMSN1 mRNA and clinicopathological parameters in 175
patients. |
Table II.
Association between expression level
of SAMSN1 mRNA and clinicopathological parameters in 175
patients.
| Variables | Low SAMSN1
mRNA in GC tissue, n | High SAMSN1
mRNA in GC tissue, n | P-value |
|---|
| Age, years |
|
| 0.710 |
|
<65 | 38 | 40 |
|
|
≥65 | 50 | 47 |
|
| Gender |
|
| 0.891 |
|
Male | 67 | 67 |
|
|
Female | 21 | 20 |
|
| Carcinoembryonic
antigen, ng/ml |
|
| 0.352 |
| ≤5 | 69 | 73 |
|
|
>5 | 19 | 14 |
|
| Carbohydrate
antigen 19-9, IU/ml |
|
| 0.467 |
|
≤37 | 69 | 72 |
|
|
>37 | 19 | 15 |
|
| Tumor location |
|
| 0.719 |
|
Entire | 6 | 8 |
|
| Upper
third | 17 | 19 |
|
| Middle
third | 31 | 24 |
|
| Lower
third | 34 | 36 |
|
| Tumor size, mm |
|
| 0.026a |
|
<60 | 43 | 57 |
|
|
≥60 | 45 | 30 |
|
| Tumor depth, UICC
classification |
|
| 0.405 |
|
pT1-3 | 42 | 47 |
|
|
pT4 | 46 | 40 |
|
|
Differentiation |
|
| 0.476 |
|
Differentiated | 37 | 32 |
|
|
Undifferentiated | 51 | 55 |
|
| Lymphatic
involvement |
|
| 0.509 |
|
Absent | 12 | 15 |
|
|
Present | 76 | 72 |
|
| Vessel
invasion |
|
| 0.708 |
|
Absent | 40 | 42 |
|
|
Present | 48 | 45 |
|
| Infiltrative growth
type |
|
| 0.598 |
|
Invasive | 31 | 34 |
|
|
Expansive | 57 | 53 |
|
| Lymph node
metastasis |
|
| 0.318 |
|
Absent | 29 | 35 |
|
|
Present | 59 | 52 |
|
| Peritoneal lavage
cytology |
|
| 0.621 |
|
Negative | 66 | 68 |
|
|
Positive | 22 | 19 |
|
| UICC stage |
|
| 0.639 |
| I | 19 | 20 |
|
| II | 14 | 15 |
|
|
III | 23 | 28 |
|
| IV | 32 | 24 |
|
 | Table III.Univariate and multivariate analysis
of prognostic factors for overall survival in 175 patients. |
Table III.
Univariate and multivariate analysis
of prognostic factors for overall survival in 175 patients.
|
|
| Univariate | Multivariate |
|---|
|
|
|
|
|
|---|
| Variables | n | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age, years
(≥65) | 97 | 1.00 | 0.63–1.60 | 0.991 | 1.08 | 0.64–1.85 | 0.782 |
| Gender
(female) | 41 | 1.14 | 0.66–1.88 | 0.638 | 1.09 | 0.58–1.97 | 0.786 |
| Carcinoembryonic
antigen (>5 ng/ml) | 33 | 1.66 | 0.93–2.79 | 0.083 | 1.13 | 0.61–2.01 | 0.688 |
| Carbohydrate
antigen 19-9 (>37 IU/ml) | 34 | 2.16 | 1.25–3.60 | 0.007 | 1.58 | 0.89–2.79 | 0.133 |
| Tumor location
(lower third) | 70 | 0.62 | 0.37–0.99 | 0.049 | 0.66 | 0.38–1.11 | 0.119 |
| Tumor size (≥60
mm) | 75 | 2.86 | 1.79–4.64 | <0.001 | 1.53 | 0.91–2.61 | 0.106 |
| Tumor depth (pT4,
UICC classification) | 86 | 3.92 | 2.39–6.65 | <0.001 | 1.72 | 0.94–3.22 | 0.079 |
| Tumor
differentiation (undifferentiated) | 106 | 1.75 | 1.08–2.92 | 0.023 | 1.22 | 0.68–2.23 | 0.507 |
| Lymphatic
involvement | 148 | 5.93 | 2.21–24.3 | <0.001 | 1.27 | 0.37–5.88 | 0.726 |
| Vessel
invasion | 93 | 2.40 | 1.48–4.00 | <0.001 | 1.70 | 1.00–3.01 | 0.049a |
| Invasive
growth | 65 | 2.64 | 1.67–4.21 | <0.001 | 1.03 | 0.55–1.96 | 0.927 |
| Lymph node
metastasis | 111 | 7.05 | 3.58–16.0 | <0.001 | 2.53 | 1.09–6.68 | 0.030a |
| Peritoneal lavage
cytology (positive) | 41 | 4.67 | 2.89–7.48 | <0.001 | 2.43 | 1.35–4.41 | 0.003a |
| Low SAMSN1
mRNA in GC tissues | 87 | 2.00 | 1.25–3.24 | 0.004 | 1.80 | 1.07–3.05 | 0.025a |
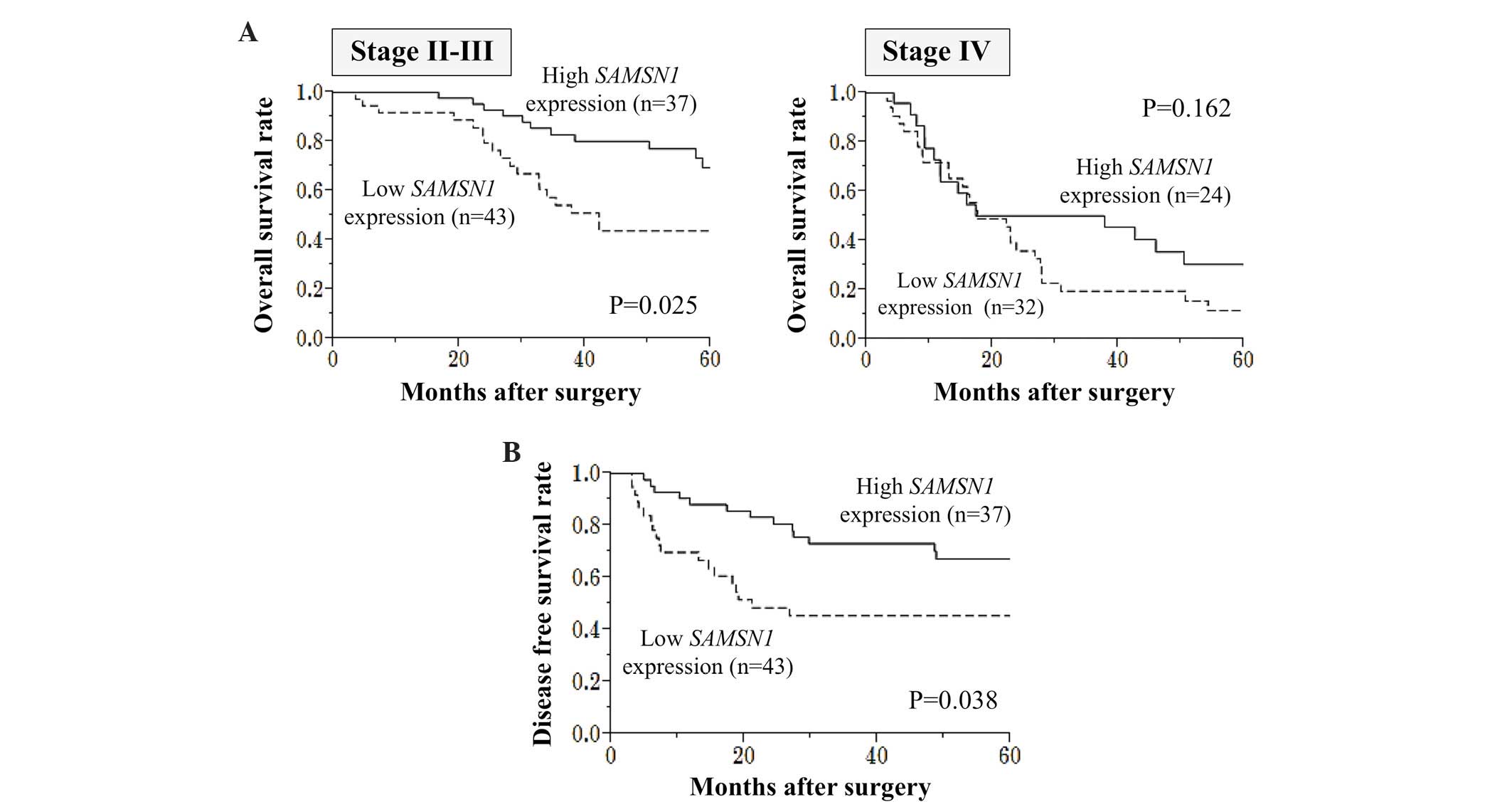
Subsequently, a subgroup analysis of patients
categorized according to UICC stage was performed. The survival
difference between the low and high SAMSN1 expression groups
was more apparent in patients with stage II/III GC (P=0.025_ than
those with stage IV GC (P=0.162) (Fig.
4A). Among 80 patients with stage II/III GC who underwent
curative surgery, those who had a low level of SAMSN1 mRNA
expression in GC tissues were more likely to have shorter disease
free survival times than those who had high SAMSN1 mRNA
expression (2-year survival rates were 50% and 81% for the low and
high SAMSN1 expression groups, respectively; P=0.038; Fig. 4B).
Discussion
The mechanism by which SAMSN1 contributes to
the tumorigenesis of digestive cancers remains to be fully
elucidated. However, it may be hypothesized that, as a B-cell
mediator, SAMSN1 may have a specific role in the initiation
and progression of GC, as this disease frequently develops from
chronically inflamed gastric mucosa, including that associated with
Helicobacter pylori-related chronic gastritis and atrophic
gastritis (28–30). Consequently, the present study sought
to investigate the status and mechanism of regulation of
SAMSN1 expression in GC. It was demonstrated that the
promoter region of SAMSN1 is methylated in a number of GC
cell lines in which SAMSN1 mRNA expression is reduced, and
that SAMSN1 expression may be restored following DNA
demethylation, despite the absence of CpG islands around the
promoter region of SAMSN1. In general, the majority of tumor
suppressor genes are suppressed through the aberrant
hypermethylation of promoter regions that contain CpG islands
(31,32). Noll et al (15) investigated the methylation status of
the SAMSN1 gene, upstream and downstream of the promoter
region, and observed that hypermethylation was associated with
suppressed expression of SAMSN1 mRNA. Given this, the
present study conducted bisulfite sequencing analysis upstream and
downstream of the SAMSN1 promoter region and observed no
methylation in GC cell lines. Further study is required to clarify
the alterative underlying molecular pathway suppressing
SAMSN1 transcription in GC.
Immunohistochemical staining and RT-qPCR analysis
revealed a direct correlation between SAMSN1 protein and
SAMSN1 mRNA expression. These findings suggest that changes
in the level of SAMSN1 mRNA are functionally significant
and, therefore, that RT-qPCR may provide a useful tool for the
quantitative analysis of SAMSN1 expression in clinical
samples (33,34).
SAMSN1 mRNA expression was significantly
downregulated in GC tissues when compared with corresponding
non-cancerous gastric tissues, and low expression of SAMSN1
mRNA was associated with more aggressive phenotypes, including
larger tumor size and shorter survival time. Furthermore,
multivariate analysis identified low SAMSN1 expression as an
independent prognostic factor. These results indicate that
SAMSN1 may function as a suppressor of GC and that
suppression of SAMSN1 expression may serve as a prognostic
indicator of this disease. Previously, it has been reported that
differences in the genetic background of tumors are reflected in
the histology, morphology and location of GCs (9,35,36). In the present study, it was observed
that SAMSN1 expression was independent of tumor location and
differentiation, indicating SAMSN1 has a similar role in all
types of GC.
The physiological function of SAMSN1 remains
to be fully elucidated. SAMSN1 is primarily expressed in
human immune tissues as well as in cell lines and primary cells
derived from patients with acute myeloid leukemia and multiple
myeloma (15,37). In addition, SAMSN1 expression
is upregulated by signaling factors that promote the activation and
differentiation of B-cells (11,13). The
present study hypothesized that chronic inflammation is caused by
H. pylori infection-induced dysregulation of immune function
and aberrant expression of SAMSN1 (38,39).
However, this hypothesis is not fully supported by the present
findings, as detailed information regarding H. pylori
infection was not collected. To develop a detailed understanding of
the oncological functions of SAMSN1, further functional
studies are required. For example, studies that aim to identify the
binding partners of SAMSN1 or those that can take advantage
of mouse models of GC to evaluate the effects of the presence or
absence of SAMSN1 on premalignant and malignant phenotypes
would be of great value in advancing our understanding of the role
of this tumor suppressor in GC (40).
There is great variability in the outcome for
patients with stage II/III GC: Certain patients respond well to
therapy and demonstrate long-term survival, while others are prone
to locoregional or distant recurrence, even following complete
curative resection (5,41). Therefore, there is a great need for
the risk stratification of stage II/III GC patients to facilitate
the appropriate management of this disease. A significant finding
from the present study was that the association between
SAMSN1 mRNA levels and postoperative prognosis for patients
with stage II/III GC was stronger than that for patients with stage
I or IV disease. This suggests that analysis of SAMSN1
expression may provide a promising tool for the identification of
stage II/III GC patients who are vulnerable to recurrence and
subsequent poor prognosis.
Taken together, the results of the present study
indicate that analysis of SAMSN1 expression may be applied
to the management of GC. The expression levels of SAMSN1 in
biopsies taken during an endoscopy or from surgically resected
tissues may be used to stratify patient risk, providing an
indication of the likelihood of recurrence and subsequent adverse
prognosis, as well as establishing a criterion for determining an
appropriate therapeutic strategy.
References
|
1
|
Kanda M, Shimizu D, Fujii T, Sueoka S,
Tanaka Y, Ezaka K, Takami H, Tanaka H, Hashimoto R, Iwata N, et al:
Function and diagnostic value of Anosmin-1 in gastric cancer
progression. Int J Cancer. 138:721–730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Songun I, Putter H, Kranenbarg EM, Sasako
M and van de Velde CJ: Surgical treatment of gastric cancer:
15-year follow-up results of the randomised nationwide Dutch D1D2
trial. Lancet Oncol. 11:439–449. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kanda M, Kobayashi D, Tanaka C, Iwata N,
Yamada S, Fujii T, Nakayama G, Sugimoto H, Koike M, Nomoto S, et
al: Adverse prognostic impact of perioperative allogeneic
transfusion on patients with stage II/III gastric cancer. Gastric
Cancer. 19:255–263. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
GASTRIC (Global Advanced/Adjuvant Stomach
Tumor Research International Collaboration) Group, ; Paoletti X,
Oba K, Burzykowski T, Michiels S, Ohashi Y, Pignon JP, Rougier P,
Sakamoto J, Sargent D, et al: Benefit of adjuvant chemotherapy for
resectable gastric cancer: A meta-analysis. JAMA. 303:1729–1737.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh
KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U, et al: Asia
Pacific Working Group on Gastric Cancer: Screening for gastric
cancer in Asia: Current evidence and practice. Lancet Oncol.
9:279–287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bang YJ, Van Cutsem E, Feyereislova A,
Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T,
et al: ToGA Trial Investigators: Trastuzumab in combination with
chemotherapy versus chemotherapy alone for treatment of
HER2-positive advanced gastric or gastro-oesophageal junction
cancer (ToGA): A phase 3, open-label, randomised controlled trial.
Lancet. 376:687–697. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kanda M, Oya H, Nomoto S, Takami H,
Shimizu D, Hashimoto R, Sueoka S, Kobayashi D, Tanaka C, Yamada S,
et al: Diversity of clinical implication of B-cell translocation
gene 1 expression by histopathologic and anatomic subtypes of
gastric cancer. Dig Dis Sci. 60:1256–1264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shah MA, Khanin R, Tang L, Janjigian YY,
Klimstra DS, Gerdes H and Kelsen DP: Molecular classification of
gastric cancer: A new paradigm. Clin Cancer Res. 17:2693–2701.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanda M, Shimizu D, Tanaka H, Shibata M,
Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, Fujii T, et
al: Metastatic pathway-specific transcriptome analysis identifies
MFSD4 as a putative tumor suppressor and biomarker for hepatic
metastasis in patients with gastric cancer. Oncotarget.
7:13667–13679. 2016.PubMed/NCBI
|
|
11
|
Zhu YX, Benn S, Li ZH, Wei E, Masih-Khan
E, Trieu Y, Bali M, McGlade CJ, Claudio JO and Stewart AK: The
SH3-SAM adaptor HACS1 is up-regulated in B cell activation
signaling cascades. J Exp Med. 200:737–747. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan Y, Zhang L, Xu T, Zhou J, Qin R, Chen
C, Zou Y, Fu D, Hu G, Chen J and Lu Y: SAMSN1 is highly expressed
and associated with a poor survival in glioblastoma multiforme.
PLoS One. 8:e819052013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Claudio JO, Zhu YX, Benn SJ, Shukla AH,
McGlade CJ, Falcioni N and Stewart AK: HACS1 encodes a novel
SH3-SAM adaptor protein differentially expressed in normal and
malignant hematopoietic cells. Oncogene. 20:5373–5377. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada H, Yanagisawa K, Tokumaru S,
Taguchi A, Nimura Y, Osada H, Nagino M and Takahashi T: Detailed
characterization of a homozygously deleted region corresponding to
a candidate tumor suppressor locus at 21q11-21 in human lung
cancer. Genes Chromosomes Cancer. 47:810–818. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Noll JE, Hewett DR, Williams SA, Vandyke
K, Kok C, To LB and Zannettino AC: SAMSN1 is a tumor suppressor
gene in multiple myeloma. Neoplasia. 16:572–585. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanda M, Nomoto S, Okamura Y, Nishikawa Y,
Sugimoto H, Kanazumi N, Takeda S and Nakao A: Detection of
metallothionein 1G as a methylated tumor suppressor gene in human
hepatocellular carcinoma using a novel method of double combination
array analysis. Int J Oncol. 35:477–483. 2009.PubMed/NCBI
|
|
17
|
Kanda M, Nomoto S, Okamura Y, Hayashi M,
Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S and Nakao A:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanda M, Nomoto S, Oya H, Takami H,
Shimizu D, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: The expression of melanoma-associated Antigen D2 both in
surgically resected and serum samples serves as clinically
rrelevant biomarker of gastric cancer progression. Ann Surg Oncol.
23:(Suppl 2). S214–S221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sakuramoto S, Sasako M, Yamaguchi T,
Kinoshita T, Fujii M, Nashimoto A, Furukawa H, Nakajima T, Ohashi
Y, Imamura H, et al: Adjuvant chemotherapy for gastric cancer with
S-1, an oral fluoropyrimidine. N Engl J Med. 357:1810–1820. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sasako M, Sakuramoto S, Katai H, Kinoshita
T, Furukawa H, Yamaguchi T, Nashimoto A, Fujii M, Nakajima T and
Ohashi Y: Five-year outcomes of a randomized phase III trial
comparing adjuvant chemotherapy with S-1 versus surgery alone in
stage II or III gastric cancer. J Clin Oncol. 29:4387–4393. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Japanese Gastric Cancer Association, .
Japanese gastric cancer treatment guidelines 2010 (ver. 3). Gastric
Cancer. 14:113–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kanda M, Sugimoto H, Nomoto S, Oya H,
Hibino S, Shimizu D, Takami H, Hashimoto R, Okamura Y, Yamada S, et
al: B-cell translocation gene 1 serves as a novel prognostic
indicator of hepatocellular carcinoma. Int J Oncol. 46:641–648.
2015.PubMed/NCBI
|
|
23
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International Union Against CancerTNM Classification of
Malignant Tumors. 7th. Wiley-Blackwell; New York, NY: pp. 73–77.
2009
|
|
24
|
Kanda M, Sugimoto H, Nomoto S, Oya H,
Shimizu D, Takami H, Hashimoto R, Sonohara F, Okamura Y, Yamada S,
et al: Clinical utility of PDSS2 expression to stratify patients at
risk for recurrence of hepatocellular carcinoma. Int J Oncol.
45:2005–2012. 2014.PubMed/NCBI
|
|
25
|
Kanda M, Nomoto S, Oya H, Takami H, Hibino
S, Hishida M, Suenaga M, Yamada S, Inokawa Y, Nishikawa Y, et al:
Downregulation of DENND2D by promoter hypermethylation is
associated with early recurrence of hepatocellular carcinoma. Int J
Oncol. 44:44–52. 2014.PubMed/NCBI
|
|
26
|
Oya H, Kanda M, Sugimoto H, Shimizu D,
Takami H, Hibino S, Hashimoto R, Okamura Y, Yamada S, Fujii T, et
al: Dihydropyrimidinase-like 3 is a putative hepatocellular
carcinoma tumor suppressor. J Gastroenterol. 50:590–600. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shimizu D, Kanda M, Nomoto S, Oya H,
Takami H, Hibino S, Suenaga M, Inokawa Y, Hishida M, Takano N, et
al: Identification of intragenic methylation in the TUSC1 gene as a
novel prognostic marker of hepatocellular carcinoma. Oncol Rep.
31:1305–1313. 2014.PubMed/NCBI
|
|
28
|
Resende C, Thiel A, Machado JC and
Ristimäki A: Gastric cancer: Basic aspects. Helicobacter. 16:(Suppl
1). S38–S44. 2011. View Article : Google Scholar
|
|
29
|
Yasui W, Sentani K, Sakamoto N, Anami K,
Naito Y and Oue N: Molecular pathology of gastric cancer: Research
and practice. Pathol Res Pract. 207:608–612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Janjigian YY and Kelsen DP: Genomic
dysregulation in gastric tumors. J Surg Oncol. 107:237–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oya H, Kanda M, Takami H, Hibino S,
Shimizu D, Niwa Y, Koike M, Nomoto S, Yamada S, Nishikawa Y, et al:
Overexpression of melanoma-associated antigen D4 is an independent
prognostic factor in squamous cell carcinoma of the esophagus. Dis
Esophagus. 28:188–195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hibino S, Kanda M, Oya H, Takami H,
Shimizu D, Nomoto S, Hishida M, Niwa Y, Koike M, Yamada S, et al:
Reduced expression of DENND2D through promoter hypermethylation is
an adverse prognostic factor in squamous cell carcinoma of the
esophagus. Oncol Rep. 31:693–700. 2014.PubMed/NCBI
|
|
35
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kanda M, Nomoto S, Oya H, Hashimoto R,
Takami H, Shimizu D, Sonohara F, Kobayashi D, Tanaka C, Yamada S,
et al: Decreased expression of prenyl diphosphate synthase subunit
2 correlates with reduced survival of patients with gastric cancer.
J Exp Clin Cancer Res. 33:882014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang D, Stewart AK, Zhuang L, Zhu Y, Wang
Y, Shi C, Keating A, Slutsky A, Zhang H and Wen XY: Enhanced
adaptive immunity in mice lacking the immunoinhibitory adaptor
Hacs1. FASEB J. 24:947–956. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lang PA, Recher M, Häussinger D and Lang
KS: Genes determining the course of virus persistence in the liver:
Lessons from murine infection with lymphocytic choriomeningitis
virus. Cell Physiol Biochem. 26:263–272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stagg J and Galipeau J: Mechanisms of
immune modulation by mesenchymal stromal cells and clinical
translation. Curr Mol Med. 13:856–867. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ringelhan M, Reisinger F, Yuan D, Weber A
and Heikenwalder M: Modeling human liver cancer heterogeneity:
Virally induced transgenic models and mouse genetic models of
chronic liver inflammation. Curr Protoc Pharmacol.
67:14.31.11–14.31.17. 2014.
|
|
41
|
Dicken BJ, Bigam DL, Cass C, Mackey JR,
Joy AA and Hamilton SM: Gastric adenocarcinoma: Review and
considerations for future directions. Ann Surg. 241:27–39.
2005.PubMed/NCBI
|