Introduction
Colorectal cancer (CRC) is one of the most
frequently diagnosed cancers and is the fourth most common cancer
diagnosed in males and the third in females (1). In China, CRC has become the fifth most
common malignancy and its incidence has shown an evident increase
over the previous decade (2). The
majority of cases of CRC are diagnosed at an advanced stage of
disease, due to inconvenient methods of current CRC screening
tests. Colonoscopy has been promoted for the detection of malignant
lesions, but the requirements of bowel preparation and the invasive
nature of colonoscopy have impeded its widespread application as a
screening tool (3). Therefore, there
is a requirement for noninvasive biomarkers to supplement and
advance current diagnostic and prognostic tools in CRC.
MicroRNAs (miRNAs) are small (23–25 nucleotides in
length) endogenous non-coding RNAs that negatively regulate gene
expression at the post-transcriptional level through RNA
interference (4,5). miRNAs are essential for various
biological processes associated with carcinogenesis, such as
proliferation, apoptosis, metastasis and differentiation (6–8). Previous
studies have demonstrated that miRNAs are abnormally expressed in
tumors and contribute to the initiation and progression of CRC as
oncogenes or tumor suppressors (9–11).
Furthermore, miRNAs can be detected in tissues and blood samples,
which can be easily obtained from individuals. Mitchell et
al (11) showed that miRNAs are
present in human plasma in a stable form that is protected from
endogenous RNase activity. Numerous studies have been focused on
cancer-derived miRNAs in the circulation system from cancer
patients (13–15), which suggests that plasma miRNAs are
novel noninvasive biomarkers for the diagnosis and prognosis of
cancers.
Among miRNAs, miRNA-23b (miR-23b) is a member of the
miR-23b/27b/24 cluster (9q22.32) and has been described as an
epigenetically silenced gene in prostate cancer (16). Functionally, overexpression of miR-23b
significantly inhibited migration, proliferation, invasion and
tumor growth (17–19). In accordance with its tumor suppressor
role, miR-23b has been found to be downregulated in numerous
cancers (20–24). However, data concerning the
association between the expression of miR-23b in plasma and CRC
were not reported yet. Therefore, to the best of our knowledge, the
present study provides the first evidence for epigenetic regulation
of miR-23b in CRC, and the expression levels of miR-23b in the
plasma of patients with CRC.
In the present study, the expression of miR-23b in
CRC cell lines and paired CRC tissues was validated, and the
methylation status of miR-23b in CRC cells was assessed.
Subsequently, the expression levels of plasma miR-23b were
evaluated in CRC patients and healthy individuals to determine
whether there was an association between the expression of miR-23b
and the clinical outcomes of CRC patients.
Materials and methods
Patients and samples
A total of 96 blood samples were obtained from
patients that underwent tumor resection for CRC between January
2009 and February 2010 at the Departments of Oncological Surgery
and Gastroenterology at the Central Hospital of Xuzhou (Xuzhou,
China). All patients did not receive anticancer intervention,
including chemotherapy or radiotherapy, prior to surgical
resection. No participants had a history of cancer. For the control
group, blood samples from 48 healthy individuals with no colonic
disease were obtained at the hospital. The healthy individuals were
matched to the CRC patients according to age and gender. In total,
20 tissue specimens (10 malignant tissues and 10 adjacent
non-malignant tissues) were snap-frozen at the time of surgery and
stored at −80°C until use. All patient samples were checked
microscopically for the presence of malignant tissue. Adjacent
non-malignant samples were obtained from tissue that appeared
morphologically normal. Clinicopathological data were collected
from the medical records of patients. The tumor-node-metastasis
(TNM) stage was assessed according to the criteria of the American
Joint Committee on Cancer (25). The
experimental protocol was approved by the Clinical Research Ethics
Committee of the Central Hospital of Xuzhou, and written informed
consent was obtained from all patients.
Cell culture and 5-aza-2-deoxycytidine
(5-Aza) treatment
In total, 5 human colon cell lines, consisting of
the LoVo, SW480, HCT-116, HT-29 and Caco-2 cell lines, were
purchased from the American Type Culture Collection (Manassas, VA,
USA) and were incubated in 5% CO2 at 37°C. LoVo cells
were cultured in Ham's F12-K medium, SW480 cells were cultured in
RPMI-1640 medium, Caco-2 cells were cultured in minimal essential
medium, and HCT-116 cells and HT-29 cells were cultured in McCoy's
5A medium. All media were purchased from Sigma-Aldrich (Merck
Millipore, Darmstadt, Germany) and supplemented with 10% fetal
bovine serum (Thermo Fisher Scientific, Inc.). The demethylation
treatment was performed with a commonly used concentration (5 µM)
of 5-Aza (Sigma-Aldrich; Merck Millipore). The fresh medium
containing 5-Aza was changed every 24 h for 3 days and treated
cells were harvested at 72 h.
Plasma preparation and RNA
extraction
Plasma samples were obtained by centrifuging the
peripheral blood (3 ml; collected in EDTA-K2 anti-coagulant tubes)
at 1,400 × g for 20 min and at 13,400 × g for 10 min
at 4°C. The plasma samples were aliquoted and stored in clean tubes
at −80°C for further analysis.
Total RNA (including miRNA) from tissues or cells or
plasma samples was extracted using Qiagen miRNeasy Mini kit
(Qiagen, Inc., Valencia, CA, USA) and Ribopure Blood RNA Isolation
kit (Ambion; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. The concentration and quality of the
isolated RNA was assessed on a NanoDrop ND-1000 Spectrophotometer
(NanoDrop, Wilmington, DE, USA). RNA samples were stored at −80°C
until use.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
miR-23b was reverse transcribed using the TaqMan
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). RT-qPCR was performed using SYBR Green PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) on
an ABI 7300HT PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Reactions were performed in a total volume of 20
µl consisting of 2 µl template (200 ng), 10 µl 2X SYBR Green Mix,
0.6 µl 200 nM forward and reverse primers, and 6 µl nuclease-free
water. The primers were used as follows: 5′-GAGCATCACATTGCCAGGG-3′
(mir-23b forward), 5′-GTGCAGGGTCCGAGGT-3′ (mir-23b reverse),
5′-CTCGCTTCGGCAGCACATAT-3′ (U6 forward) and
5′-TTGCGTGTCATCCTTGCG-3′ (U6 reverse). All primers were purchased
from Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
The PCR conditions were as follows: Initial denaturation at 95°C
for 5 min, followed by 45 cycles of 95°C for 30 sec, 56°C for 20
sec, and 72°C for 30 sec. RNU6B (U6) was used as an internal
control. The relative expression levels of miR-23b were normalized
to the internal control U6, and were calculated using the
2−ΔΔCq method (ΔCq = CqmiR-23b -
CqU6) (26). Each
experiment was conducted in triplicate.
DNA extraction, bisulfite conversion
and methylation-specific PCR (MSP)
DNA was isolated from tissue samples and CRC cells
using DNeasy Blood & Tissue kit (Qiagen, Inc.). The extracted
DNA was then subjected to bisulfite treatment, which was performed
using the EpiTect Bisulfite kit (Qiagen, Inc.), according to the
manufacturer's instructions. The amplification reaction was
conducted under the following conditions: 95°C for 5 min, then 46
cycles of 95°C for 45 sec, 56°C for 35 sec, 72°C for 50 sec, and
finally 10 min at 72°C. The PCR products were visualized on 2%
agarose gels.
Statistical analysis
The SPSS 16.0 software (SPSS, Inc., Chicago, IL,
USA) and Prism (GraphPad Software, La Jolla, CA, USA) was used to
perform statistical analysis of results. The Wilcoxon t-test
was used to compare the paired samples obtained from primary CRC
tissues and adjacent non-cancerous colorectal tissues. The
Mann-Whitney test was performed to compare the expression levels of
miR-23b in plasma from CRC patients and healthy control
individuals. The diagnostic performance of plasma miR-23b was
assessed by receiver operating characteristic (ROC) curve analysis.
The cut-off value (median of the expression level) was used to
define the mir-23b expression status (low or high mir-23b). The
association between the expression status of mir-23b and
clinicopathological characteristics were determined by applying
Fisher's exact test or χ2 test, as appropriate. The
overall survival time was determined as the time between the date
of surgery and mortality, and was calculated using the Kaplan-Meier
method. Differences between groups were compared using Log-rank
test. Cox proportional hazard regression was used to estimate the
univariate and multivariate analysis, and calculate hazard ratios
(HRs) and 95% confidence intervals (CIs). All tests were
two-tailed, and P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-23b in primary CRC
tissues and CRC cells
To verify previous studies (24,27) that
reported downregulation of miR-23b in primary CRC, RT-qPCR was used
to evaluate miR-23b expression in 10 paired CRC tissues and
adjacent non-cancerous tissues. Additionally, the expression levels
of miR-23b in 5 human CRC cell lines, consisting of the LoVo,
SW480, HCT-116, HT-29 and Caco-2 cell lines, were also analyzed by
RT-qPCR. As shown in Fig. 1A, the
miR-23b expression levels were significantly downregulated in CRC
tissues compared with non-cancerous tissues (0.016±0.004 vs.
0.086±0.019; P<0.001). The miR-23b expression levels were also
significantly decreased in CRC cells compared with non-cancerous
tissues (0.010±0.001 vs. 0.086±0.019; P<0.001; Fig. 1B).
Methylation status of miR-23b in
CRC
Since previous studies have described that miR-23b
is an epigenetic target in prostate cancer and glioma stem cells
(16,28), the present study examined the
methylation status of the miR-23b gene in the LoVo, SW480, HCT-116,
HT-29 and Caco-2 cell lines, and two non-malignant colorectal
tissue samples. As shown in Fig. 2A,
the promoter region of the miR-23b gene was highly methylated in
all 5 CRC cell lines, while it was highly unmethylated in
non-malignant colorectal tissues. Furthermore, treatment with 5-Aza
markedly increased miR-23b expression (Fig. 2B) in the CRC cell lines. These results
suggest that promoter methylation has an important role in the
downregulation of miR-23b in CRC.
Expression of miR-23b in the plasma of
patients with CRC
Following validation of miR-23b expression in paired
CRC tissues, the plasma expression level of miR-23b in 96 patients
with CRC and 48 healthy control individuals was measured. The
relative miR-23b expression level in plasma from patients with CRC
was 0.010±0.009, which was significantly decreased compared with
the healthy control individuals (0.053±0.041; P<0.001; Fig. 3A).
To determine whether miR-23b expression can
discriminate between patients with CRC and healthy control
individuals, ROC analysis was performed. The optimal cut-off value
for sensitivity and specificity was determined based on the highest
Youden's Index in ROC curve analysis. The area under the ROC curve
(AUC) of 0.842 (sensitivity, 84.38%; specificity, 77.08%; 95% CI,
0.763–0.922) indicated that miR-23b expression can discriminate
between patients with CRC and healthy control individuals (Fig. 3B).
Association between the plasma miR-23b
levels and the clinicopathological variables
The median value (0.007) of miR-23b expression in
all CRC plasma samples, measured by RT-qPCR, was used as a
threshold point to classify 96 patients into high- and low-miR-23b
expression groups. The association between the plasma miR-23b
expression level and the clinicopathological parameters of the
patients with CRC is shown in Table
I. The present results revealed that the miR-23b expression
levels in the plasma of patients with CRC were not significantly
associated with gender, age, histological type, lymph node
metastasis or tumor location. The associations between the miR-23b
expression level in the plasma and TNM stage (P=0.007), tumor depth
(P=0.041), distant metastasis (P<0.001) and recurrence
(P<0.001) were all statistically significant.
 | Table I.Association between plasma miR-23b
expression and clinicopathological variables in patients with
colorectal cancer. |
Table I.
Association between plasma miR-23b
expression and clinicopathological variables in patients with
colorectal cancer.
|
|
| miR-23b expression,
n |
|
|---|
|
|
|
|
|
|---|
| Variables | Total, n | High | Low | P-value |
|---|
| Patient number | 96 | 48 | 48 |
|
| Gender |
|
|
|
0.826 |
|
Female | 29 | 16 | 13 |
|
|
Male | 67 | 32 | 35 |
|
| Age |
|
|
|
0.280 |
| <60
years | 45 | 24 | 21 |
|
| ≥60
years | 51 | 24 | 27 |
|
| Histological
type |
|
|
|
0.545 |
|
Differentiated | 47 | 22 | 25 |
|
|
Undifferentiated | 49 | 26 | 23 |
|
| TNM stage |
|
|
|
0.007 |
| I,
II | 39 | 19 | 20 |
|
| III,
IV | 57 | 29 | 28 |
|
| Tumor depth |
|
|
|
0.041 |
|
T1-2 | 46 | 21 | 25 |
|
|
T3-4 | 50 | 27 | 23 |
|
| Lymph node
metastasis |
|
|
|
0.103 |
| No | 44 | 22 | 22 |
|
|
Yes | 52 | 26 | 26 |
|
| Distant
metastasis |
|
|
|
<0.001 |
| No | 73 | 37 | 36 |
|
|
Yes | 23 | 11 | 12 |
|
| Recurrence |
|
|
|
<0.001 |
| No | 74 | 36 | 38 |
|
|
Yes | 22 | 12 | 10 |
|
| Location |
|
|
|
0.687 |
|
Rectum | 46 | 24 | 22 |
|
|
Colon | 50 | 24 | 26 |
|
Expression of plasma miR-23b and
prognosis
To determine whether miR-23b has prognostic
significance, Kaplan-Meier survival analysis and Cox proportional
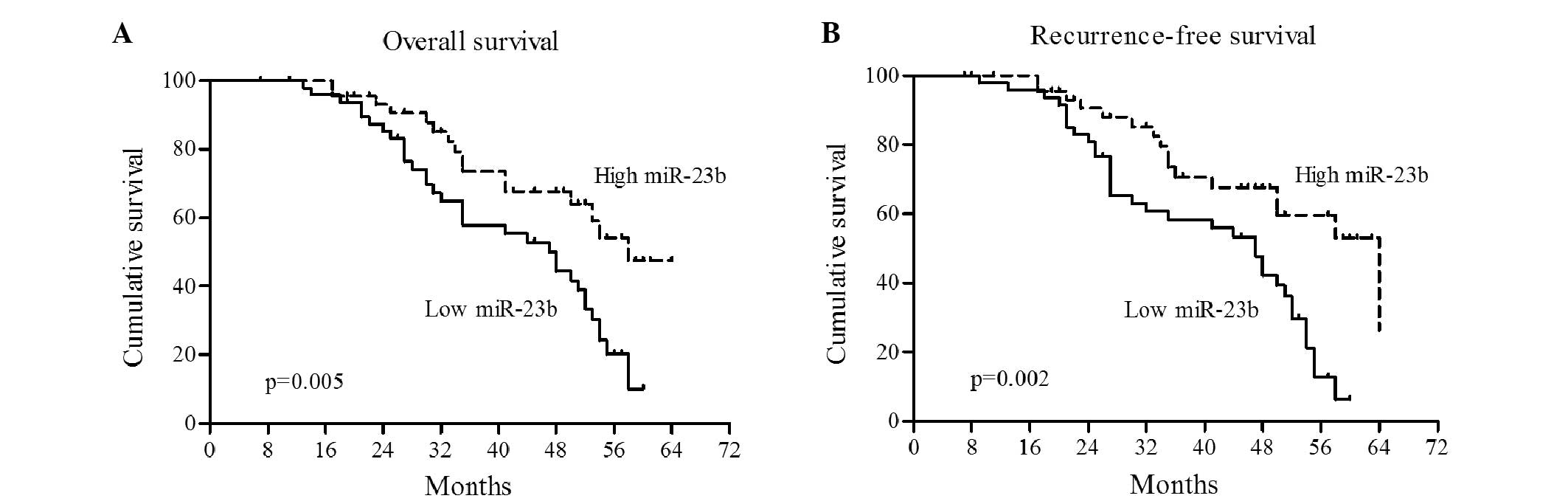
hazard analysis were performed. As shown in Fig. 4, Kaplan-Meier analysis showed that the
high miR-23b group had a significantly increased overall survival
rate compared with the low miR-23b group (P=0.005; Fig. 4A). In addition, CRC patients with a
high miR-23b level in plasma had a longer recurrence-free survival
time compared with the low miR-23b patients (P=0.002; Fig. 4B).
The association between clinicopathological
parameters and expression level of miR-23b with the clinical
outcome was also analyzed in patients with CRC. The Cox
proportional hazard regression analysis showed that of age >60
years (P=0.005), increased TNM stage (P<0.001), increased tumor
depth (P=0.001), presence of lymph node metastasis (P=0.007) and
distant metastasis (P<0.001) were significant prognositic
factors (Table II). The low
expression level of miR-23b was associated with a relative risk of
death of 2.447 (95% CI, 1.392–4.301; P=0.002).
 | Table II.Univariate and multivariate analysis
for overall survival of patients with colorectal cancer. |
Table II.
Univariate and multivariate analysis
for overall survival of patients with colorectal cancer.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Gender
(female/male) | 1.520 | 0.816–2.834 |
0.187 |
|
|
|
| Age (<60
years/≥60 years) | 2.499 | 1.312–4.763 |
0.005 |
3.305 | 1.628–6.709 |
0.001 |
| Histological type
(differentiated/undifferentiated) | 1.489 | 0.875–2.535 |
0.142 |
|
|
|
| TNM stage (I,
II/III, IV) | 3.047 | 1.630–5.697 | <0.001 |
|
|
|
| Tumor depth
(T1-2/T3-4) | 2.525 | 1.434–4.447 |
0.001 |
|
|
|
| Lymph node
metastasis (no/yes) | 2.179 | 1.237–3.839 |
0.007 |
|
|
|
| Distant metastasis
(no/yes) | 9.356 |
5.214–16.792 | <0.001 | 13.277 |
6.844–25.758 | <0.001 |
| Recurrence
(no/yes) | 1.293 | 0.721–2.318 |
0.389 |
|
|
|
| Location
(rectum/colon) | 1.396 | 0.819–2.382 |
0.220 |
|
|
|
| miR-23b expression
(low/high) | 2.447 | 1.392–4.301 |
0.002 |
|
|
|
Multivariate analysis was performed using age, TNM
stage, tumor depth, lymph node metastasis, distant metastasis and
the expression level of miR-23b. These data revealed that only age
>60 years old (HR, 3.305; 95% CI, 1.628–6.709; P=0.001) and
presence of distant metastasis (HR, 13.277; 95% CI, 6.844–25.758;
P<0.001) were independently associated with a significantly
increased risk of mortality (Table
II). The expression level of miR-23b in the plasma of CRC
patients was not a significant independent risk factor.
Discussion
In previous years, studies have demonstrated that
detecting levels of plasma miRNAs in the blood is viable for
clinical application (13–15). Measurement of tumor-derived miRNAs in
the plasma is an important approach for the blood-based detection
of human cancer (12). Numerous
studies have indicated that plasma miRNAs have potential diagnostic
or prognostic value in various types of tumors, including breast
cancer (14), leukemia (29), gastric cancer (30), oral cancer (31) and colon cancer (32).
miR-23b has been demonstrated to be involved in
several tumor-associated biological process, particularly in tumor
metastasis. For example, overexpression of miR-23b has been shown
to inhibit migration in prostate cancer cells (16,33), and
hepatocellular carcinoma cells (17).
In glioma, overexpression of miR-23b significantly inhibited cell
migration and invasion while inhibition of miR-23b expression
significantly increased migration (18). Increased expression of miR-23b also
inhibited proliferation in prostate cancer (16), epithelial ovarian cancer (19), and hepatocellular carcinoma, where it
acts as a tumor suppressor by targeting the urokinase-type
plasminogen activator and c-met (17). In addition, miR-23b functions as a
regulator for G0-G1 cell-cycle arrest (16,34).
miR-23b was also found to regulate transforming growth
factor-β/bone morphogenetic protein signaling and affected liver
stem cell differentiation (35). In
colon cancer, Zhang et al demonstrated that miR-23b is
downregulated in human colon cancer and mediates multiple steps of
metastasis, including tumor growth, invasion and angiogenesis
(24). Therefore, it was hypothesized
that quantitative measuring of miR-23b in the plasma may be a
promising biomarker in CRC patients.
Firstly, the expression level of miR-23b in CRC
cells and paired tissues from CRC patients was validated. A
significantly decreased expression level of miR-23b was found in
CRC cells and malignant tissues compared with non-malignant
tissues. Similarly, several studies have demonstrated that miR-23b
expression is significantly downregulated in various types of
tumor, including prostate, hepatocellular, bladder, endometrial
carcinosarcoma and colon cancer (16,17,20–24).
Conversely, miR-23b is overexpressed in oral squamous cell
carcinoma (36), glioma (37) and breast cancer (38,39).
Tissue specificity may be the reason of differential expression of
miR-23b in cancers. Additionally, considering promoter methylation
plays a role in the downregulation of miR-23b in prostate cancer
(16), the MSP method was applied to
analyze the methylation status of miR-23b in CRC. miR-23b was
highly methylated in CRC cells, but it was unmethylated in
non-malignant tissues. To further analyze whether methylation of
the miR-23b promoter regulates its expression, cancer cell lines
were treated with the demethylating agent 5-Aza. Subsequent to
5-Aza treatment, miR-23b expression was significantly upregulated
in all 5 cell lines. These results indicate that methylation of the
miR-23b promoter directly regulates its transcriptional repression
in CRC cells.
RT-qPCR demonstrated that miR-23b was significantly
downregulated in plasma from CRC patients compared with healthy
controls. Based on the ROC curve analysis, the plasma level of
miR-23b had a clinically satisfactory degree of specificity and
sensitivity with an AUC of 0.842, suggesting that miR-23b
expression can discriminate CRC from healthy controls and
potentially be used as a diagnostic marker for CRC. In addition to
the present study, miR-23b expression also distinguishes between
normal and bladder cancer tissues with an AUC of 0.885 (16). The diagnostic value of plasma miR-23b
for cancer requires additional study in a large trial. The present
findings revealed that the miR-23b expression in plasma was
significantly associated with TNM stage, tumor depth, distant
metastasis and tumor recurrence. Similarly, low miR-23b expression
associates with the development of metastases in breast cancer
(40) and colon cancer lung
metastasis (24). Low expression of
miR-23b in plasma was significantly associated with poor overall
survival and shorter recurrence-free survival in CRC patients.
These results were consistent with previous studies of bladder
cancer (16), ovarian cancer
(19) and prostate cancer (21). Therefore, miR-23b may potentially be a
powerful prognostic marker for cancer patients.
In conclusion, miR-23b is epigenetically
downregulated in CRC and the present data suggest a potential
diagnostic and prognostic value of plasma miR-23b levels in
discriminating between patients with malignancy and healthy
individuals, and also for predicting overall survival for patients
with CRC.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wan DS: Epidemiologic trend of and
strategies for colorectal cancer. Ai Zheng. 28:897–902. 2009.(In
Chinese). PubMed/NCBI
|
|
3
|
van Dam J and Friedman LS: Missed cancers
at colonscopy: Learning the hard way. Gastrointest Endosc.
45:530–533. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhaumik D, Scott GK, Schokrpur S, Patil
CK, Campisi J and Benz CC: Expression of microRNA-146 suppresses
NF-kappaB activity with reduction of metastatic potential in breast
cancer cells. Oncogene. 27:5643–5647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abella V, Valladares M, Rodriguez T, Haz
M, Blanco M, Tarrío N, Iglesias P, Aparicio LA and Figueroa A:
MiR-203 regulates cell proliferation through its influence on Hakai
expression. PLoS One. 7:e525682012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Huang H, Sun L, Yang M, Pan C, Chen
W, Wu D, Lin Z, Zeng C, Yao Y, et al: MiR-21 indicates poor
prognosis in tongue squamous cell carcinomas as an apoptosis
inhibitor. Clin Cancer Res. 15:3998–4008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lanza G, Ferracin M, Gafà R, Veronese A,
Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM and Negrini M:
Mrna/MicroRNAs gene expression profile in microsatellite unstable
colorectal cancer. Mol Cancer. 6:542007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Link A, Balaguer F, Shen Y, Nagasaka T,
Lozano JJ, Boland CR and Goel A: Fecal MicroRNAs as novel
biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers
Prev. 19:1766–1774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: Microrna-21 (mir-21)
post-transcriptionally downregulates tumor suppressor pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gilad S, Meiri E, Yogev Y, Benjamin S,
Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed
N, et al: Serum microRNAs are promising novel biomarkers. PLoS One.
3:e31482008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7:e297702012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, Poon TC, Ng SS and Sung JJ: Differential expression of
microRNAs in plasma of patients with colorectal cancer: A potential
marker for colorectal cancer screening. Gut. 58:1375–1381. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Majid S, Dar AA, Saini S, Arora S,
Shahryari V, Zaman MS, Chang I, Yamamura S, Tanaka Y, Deng G and
Dahiya R: MiR-23b represses proto-oncogene Src kinase and functions
as methylation-silenced tumor suppressor with diagnostic and
prognostic significance in prostate cancer. Cancer Res.
72:6435–6446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salvi A, Sabelli C, Moncini S, Venturin M,
Arici B, Riva P, Portolani N, Giulini SM, De Petro G and Barlati S:
MicroRNA-23b mediates urokinase and c-met downmodulation and a
decreased migration of human hepatocellular carcinoma cells. FEBS
J. 276:2966–2982. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Loftus JC, Ross JT, Paquette KM, Paulino
VM, Nasser S, Yang Z, Kloss J, Kim S, Berens ME and Tran NL: MiRNA
expression profiling in migrating glioblastoma cells: Regulation of
cell migration and invasion by miR- 23b via targeting of Pyk2. PLoS
One. 7:e398182012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li W, Liu Z, Chen L, Zhou L and Yao Y:
MicroRNA-23b is an independent prognostic marker and suppresses
ovarian cancer progression by targeting runt-related transcription
factor-2. FEBS Lett. 588:1608–1615. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tong AW, Fulgham P, Jay C, Chen P, Khalil
I, Liu S, Senzer N, Eklund AC, Han J and Nemunaitis J: MicroRNA
profile analysis of human prostate cancers. Cancer Gene Ther.
16:206–216. 2009.PubMed/NCBI
|
|
21
|
Goto Y, Kojima S, Nishikawa R, Enokida H,
Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T and Seki
N: The microRNA-23b/27b/24-1 cluster is a disease progression
marker and tumor suppressor in prostate cancer. Oncotarget.
5:7748–7759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun T, Yang M, Chen S, Balk S, Pomerantz
M, Hsieh CL, Brown M, Lee GS and Kantoff PW: The altered expression
of MiR-221/-222 and MiR-23b/-27b is associated with the development
of human castration resistant prostate cancer. Prostate.
72:1093–1103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Castilla MÁ, Moreno-Bueno G, Romero-Pérez
L, Van De Vijver K, Biscuola M, López-García MÁ, Prat J,
Matías-Guiu X, Cano A, Oliva E and Palacios J: Micro-RNA signature
of the epithelial-mesenchymal transition in endometrial
carcinosarcoma. J Pathol. 223:72–80. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin
S, Sun C, Ma M, Huang Y and Xi JJ: Genome-wide functional screening
of miR-23b as a pleiotropic modulator suppressing cancer
metastasis. Nat Commun. 2:5542011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Obrocea FL, Sajin M, Marinescu EC and
Stoica D: Colorectal cancer and the 7th revision of the TNM staging
system: Review of changes and suggestions for uniform pathologic
reporting. Rom J Morphol Embryol. 52:537–544. 2011.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou X, Xu X, Wang J, Lin J and Chen W:
dentifying miRNA/mRNA negative regulation pairs in colorectal
cancer. Sci Rep. 5:129952015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Geng J, Luo H, Pu Y, Zhou Z, Wu X, Xu W
and Yang Z: Methylation mediated silencing of miR-23b expression
and its role in glioma stem cells. Neurosci Lett. 528:185–189.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tanaka M, Oikawa K, Takanashi M, Kudo M,
Ohyashiki J, Ohyashiki K and Kuroda M: Down-regulation of miR-92 in
human plasma is a novel marker for acute leukemia patients. PLoS
One. 4:e55322009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsujiura M, Ichikawa D, Komatsu S,
Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi
K, Fujiwara H, et al: Circulating microRNAs in plasma of patients
with gastric cancers. Br J Cancer. 102:1174–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW
and Lin SC: Increase of microRNA miR-31 level in plasma could be a
potential marker of oral cancer. Oral Dis. 16:360–364. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ishteiwy RA, Ward TM, Dykxhoorn DM and
Burnstein KL: The microRNA −23b/-27b cluster suppresses the
metastatic phenotype of castration-resistant prostate cancer cells.
PLoS One. 7:e521062012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang KC, Garmire LX, Young A, Nguyen P,
Trinh A, Subramaniam S, Wang N, Shyy JY, Li YS and Chien S: Role of
microRNA-23b in flow-regulation of Rb phosphorylation and
endothelial cell growth. Proc Natl Acad Sci USA. 107:3234–3239.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rogler CE, Levoci L, Ader T, Massimi A,
Tchaikovskaya T, Norel R and Rogler LE: MicroRNA-23b cluster
microRNAs regulate transforming growth factor-beta/bone
morphogenetic protein signaling and liver stem cell differentiation
by targeting smads. Hepatology. 50:575–584. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Scapoli L, Palmieri A, Lo Muzio L,
Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M and
Carinci F: MicroRNA expression profiling of oral carcinoma
identifies new markers of tumor progression. Int J Immunopathol
Pharmacol. 23:1229–1234. 2010.PubMed/NCBI
|
|
37
|
Chen L, Han L, Zhang K, Shi Z, Zhang J,
Zhang A, Wang Y, Song Y, Li Y, Jiang T, et al: VHL regulates the
effects of miR-23b on glioma survival and invasion via suppression
of HIF-1α/VEGF and β-catenin/Tcf-4 signaling. Neuro Oncol.
14:1026–1036. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Paris O, Ferraro L, Grober OM, Ravo M, De
Filippo MR, Giurato G, Nassa G, Tarallo R, et al: Direct regulation
of microRNA biogenesis and expression by estrogen receptor beta in
hormone-responsive breast cancer. Oncogene. 31:4196–4206. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin L, Wessely O, Marcusson EG, Ivan C,
Calin GA and Alahari SK: Prooncogenic factors miR-23b and miR-27b
are regulated by Her2/Neu, EGF, and TNF-α in breast cancer. Cancer
Res. 73:2884–2896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pellegrino L, Stebbing J, Braga VM,
Frampton AE, Jacob J, Buluwela L, Jiao LR, Periyasamy M, Madsen CD,
Caley MP, et al: MiR-23b regulates cytoskeletal remodeling,
motility and metastasis by directly targeting multiple transcripts.
Nucleic Acids Res. 41:5400–5412. 2013. View Article : Google Scholar : PubMed/NCBI
|