Introduction
Colorectal cancer (CRC) is one of the most common
gastrointestinal malignancies worldwide. It is ranked fifth in
terms of cancer-associated mortality in China (1). The incidence of colon cancer has been
increasing in a large number of countries over the previous 20
years (2,3). The 5-year relative survival rate for
colon cancer patients is 64.2% (4).
Approximately 694,000 patients succumb to CRC globally every year
(5). The CRC incidence and mortality
both increased prior to the onset of organized screening in
middle-income countries, such as China and Thailand (6), which inflicts a high socio-economic
burden.
The majority of cases of colorectal cancer develop
from polyps, however, patients with colon polyps and precancerous
tumors typically have no obvious symptoms. For diagnosis of CRC,
sigmoidoscopy or colonoscopy with biopsy can be used to confirm the
presence of cancer tissues. Surgery is the most common method of
treatment for CRC. Due to advances in surgical techniques and
neoadjuvant chemotherapy, the median survival of patients with
early-stage colon cancer continues to improve (7), however, prognosis remains poor for
patients with advanced colorectal cancer. Previous studies
indicated that 10–30% of patients with CRC eventually develop
recurrent disease, despite receiving radical treatment (8,9).
The pathology and clinical stage of colon cancer
tumors has been shown to be closely associated with oncogene
activation and suppression (10). The
Wnt/β-catenin pathway is thought to be central to the process of
carcinogenesis in CRC (11–13). Recently, a novel proto-oncogene,
frequently rearranged in advanced T-cell lymphomas 1 (FRAT1), has
increasingly received attention. The FRAT1 gene has been
demonstrated to be involved in the regulation of β-catenin
expression and secretion (14).
Previous studies have demonstrated that in non-small cell lung
cancer and gastric cancer tissues, overexpression of FRAT1
activates the Wnt signaling pathway and promotes tumor malignancy
(15,16). However, few studies have investigated
the effect of FRAT1 expression in colon cancer. In the present
study, colon cancer specimens were obtained from colon cancer
patients undergoing gastrointestinal surgery. The aim of the
present study was to investigate the expression of FRAT1 in colon
cancer, and to analyze the association between FRAT1 expression and
proliferation and migration of tumor cells.
Materials and methods
Patients and tissue samples
A total of 147 colon cancer tissue samples and
adjacent normal control samples were obtained from patients
diagnosed with colon cancer who underwent radical surgery at The
Second Hospital of Shandong University (Jinan, China) between
January 2013 and June 2014. All patients exhibited primary tumors
and none of the patients had received chemotherapy or radiation
therapy prior to tumor excision. The patient cohort included 93
males and 54 females, with a median age of 62 years (range, 37–72
years). Patient clinicopathological factors are shown in Table I. The present study was approved by
the Ethics Committee of the Second Hospital of Shandong University
(Jinan, China) and additionally received institutional approval.
The experiments were performed in accordance with the World Medical
Association Declaration of Helsinki Ethical Principles for Medical
Research.
 | Table I.Clinicopathological factors of 147
colon cancer patients. |
Table I.
Clinicopathological factors of 147
colon cancer patients.
| Parameter | Patients, n |
|---|
| Gender |
|
| Male | 93 |
|
Female | 54 |
| Age (median, range)
(years) | 62 (37–72) |
| TNM stage |
|
| I | 26 |
| II | 45 |
| III | 51 |
| IV | 25 |
| Tumor
differentiation |
|
|
Well-differentiated | 41 |
|
Moderately differentiated | 82 |
| Poorly
differentiated | 24 |
Western blot analysis
Resected tissues were washed using
phosphate-buffered saline (PBS), cut into sections and lysed with
RIPA lysis and extraction buffer (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The lysates were centrifuged in an ice bath to
obtain the supernatant and total protein was calculated using the
bicinchoninic acid assay method (Pierce BCA Protein Assay kit;
Thermo Fisher Scientific, Inc.). A total of 50 µg protein was used
for each sample. Following electrophoresis with 12% gel, samples
were transferred to polyvinylidene fluoride membranes and incubated
with FRAT1 rabbit monoclonal antibody (1:500; catalog no.,
ab108405; Abcam, Cambridge, UK) overnight at 4°C. Next, the
membranes were incubated with goat anti-rabbit IgG-horseradish
peroxidase (1:2,000; catalog no., sc-2005; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 37°C for 2 h and
visualized using a chemiluminescence detection reagent (Applygen
Technologies, Inc., Beijing, China). The western blot gray values
were quantified using ImageJ software (National Institutes of
Health, Bethesda, MA, USA).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
RNA was isolated from the tissue samples using
mechanical homogenization and TRIzol reagent (Takara Biotechnology
Co., Ltd., Dalian, China). cDNA was synthesized from the total RNA
using SuperScript III RNase H-Reverse Transcriptase (Takara
Biotechnology Co., Ltd.). Gene expression was quantified using a
One-Step SYBR PrimeScript RT-PCR kit (Takara Biotechnology Co.,
Ltd.). The primers were designed as described previously (17). The sequences of the primers were as
follows: Forward, 5′-GCCCTGTCTAAAGTGTATTTTCAG-3′ and reverse,
5′-CGCTTGAGTAGGACTGCAGAG-3′ for FRAT1 (Invitrogen; Thermo Fisher
Scientific, Inc.). GADPH was used as the internal reference gene
and its primers were as follows: forward, 5′-GAAGTGAAGGTCGGAGTCA-3′
and reverse, 5′-TTCACACCCATGACGAACAT-3′. The gene expression was
analyzed on the QuantStudio™ 6 Flex Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). PCR was performed in a
10 µl reaction volume, which consisted of 5 µl One-Step SYBR buffer
III, 0.2 µl Takara Ex Taq HS, 0.2 µl PrimerScript RT enzyme
mix II, 0.2 µl forward primer, 0.2 µl reverse primer, 1 µl RNA and
3.2 µl RNase-free dH20. PCR was performed under the
following conditions: Initial denaturation at 90°C for 5 min,
followed by 40 cycles of denaturation at 95°C for 45 sec, annealing
at 55°C for 40 sec and extension at 72°C for 50 sec (18).
Cell transfection with FRAT1
The specific sequence of the shRNA targeting FRAT1
was designed as previously described (19–22) using
the following primer sequences: Forward,
5′-GCAGTTACGTGCAAAGCTTTTCAAGAGAAAGCTTTGCACGTACTGC-3′ and reverse,
3′-CGTCAATGCACGTTTCGAAAAGTTCTCTTTCGAAACGTGCATTGACG-5′. The shRNA
template was then added to two endonucleases, HindIII and
BamH1, for digestion. The synthetic template was annealed to
form a double-stranded oligo and then cloned into the vector
pLV-H1-EF1a-puro (Biosettia Inc., San Diego, CA, USA). Finally,
recombinant pshRNA-FRAT1 was successfully constructed and verified
by Sanger dideoxy DNA sequencing. HT-29 cells were transfected with
the recombinant plasmid to screen the stable cell line for FRAT1
inhibition. In addition, HT-29 cells were tranfected with empty
vector pLV-H1-EF1a-puro and used as the control. To ensure
efficient transfection, reverse transcription (RT)-PCR and western
blot analysis were used to analyze FRAT1 gene and protein
expression.
MTT assay
Cell proliferation was analyzed using the MTT
colorimetric method. HT-29 cells (1×104 cells/well) were
subcultured to 80% confluence and seeded in 96-well plates. The
cells were incubated for 48 h, and were cultured for 4 h with 5
mg/ml MTT (Sigma-Aldrich, St. Louis, MO, USA). The optical density
(OD) was then measured (ELx800 UV universal microplate reader;
Bio-Tek Instruments, Inc., Winooski, VT, USA) and the inhibition
rate was calculated according to the following formula: Inhibition
rate = 1 - experimental OD / control OD (23).
Cell migration was performed using Transwell inserts
(8-µm pore size; Merck Millipore, Darmstadt, Germany). The upper
chamber was coated with 1 mg/ml Matrigel (Corning, Inc., Corning,
NY, USA). The cells were incubated without serum for 12 h and
seeded at a density of 2×105/ml into the upper chamber,
while 600 µl 10% serum medium was placed into the lower chamber.
The cells were cultured for 36 h and subsequently the cells on the
upper surface were removed gently with a cotton swab. The Transwell
chambers were fixed in 95% ethanol for 15 min and washed with PBS 3
times. Eosin was used to stain the chambers for 10 min, followed by
washing with PBS. Finally, the chambers were viewed under a
high-powered inverted microscope (CKX41; Olympus Corp., Tokyo,
Japan). Cells were counted in 6 random fields at magnification,
×200.
Statistical analysis
All data analysis was performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Data are expressed as the
mean ± standard error of the mean. Differences between groups were
determined using Student's t-test and the correlation between FRAT1
expression, TNM stage and pathological stage was determined using
χ2 and Spearman's rank correlation analysis. P<0.05
was considered to indicate a statistically significant
difference.
Results
FRAT1 protein and gene expression in
colon cancer and normal tissues
As shown in Fig. 1,
mRNA expression of FRAT1 was significantly increased in colon
cancer tissues compared with adjacent tissues at different
pathological stages (P<0.05; Fig.
1A) and various Tumor-Node-Metastasis (TNM) stages (P<0.05;
Fig. 1B) (24). Western blot results revealed that the
expression of FRAT1 significantly increased in colon cancer tissues
of various pathological (P<0.05; Fig.
1C) and TNM stages (P<0.05; Fig.
1D) compared with adjacent tissues.
Correlation between
clinicopathological stage and FRAT1 expression
In the colon cancer tissues, increased FRAT1 gene
and protein expression was found to significantly correlate with
increased tumor malignancy and a higher TNM stage (P<0.05)
(Table II).
 | Table II.FRAT1 expression in 147 colon cancer
tissue samples. |
Table II.
FRAT1 expression in 147 colon cancer
tissue samples.
| Clinical
characteristics | Patients, n | FRAT1 expression
(±SEM) | FRAT1 WB gray
(±SEM) | P-value |
|---|
| Histological
grade |
|
|
|
|
| Well
differentiated | 41 | 7.39±2.65 | 2.42±2.21 | 0.012 |
|
Moderately differentiated | 82 | 4.39±1.98 | 4.92±1.77 |
| Poor
differentiated | 24 | 3.12±1.66 | 5.78±1.98 |
|
| TNM stage |
|
|
| 0.015 |
| I | 26 | 2.13±0.79 | 1.79±0.32 |
| II | 45 | 3.96±1.46 | 2.12±0.51 |
| III | 51 | 5.77±1.82 | 3.79±0.98 |
| IV | 25 | 8.02±2.04 | 5.13±1.22 |
Effects of regulating FRAT1 expression
on colon cancer cell proliferation and apoptosis
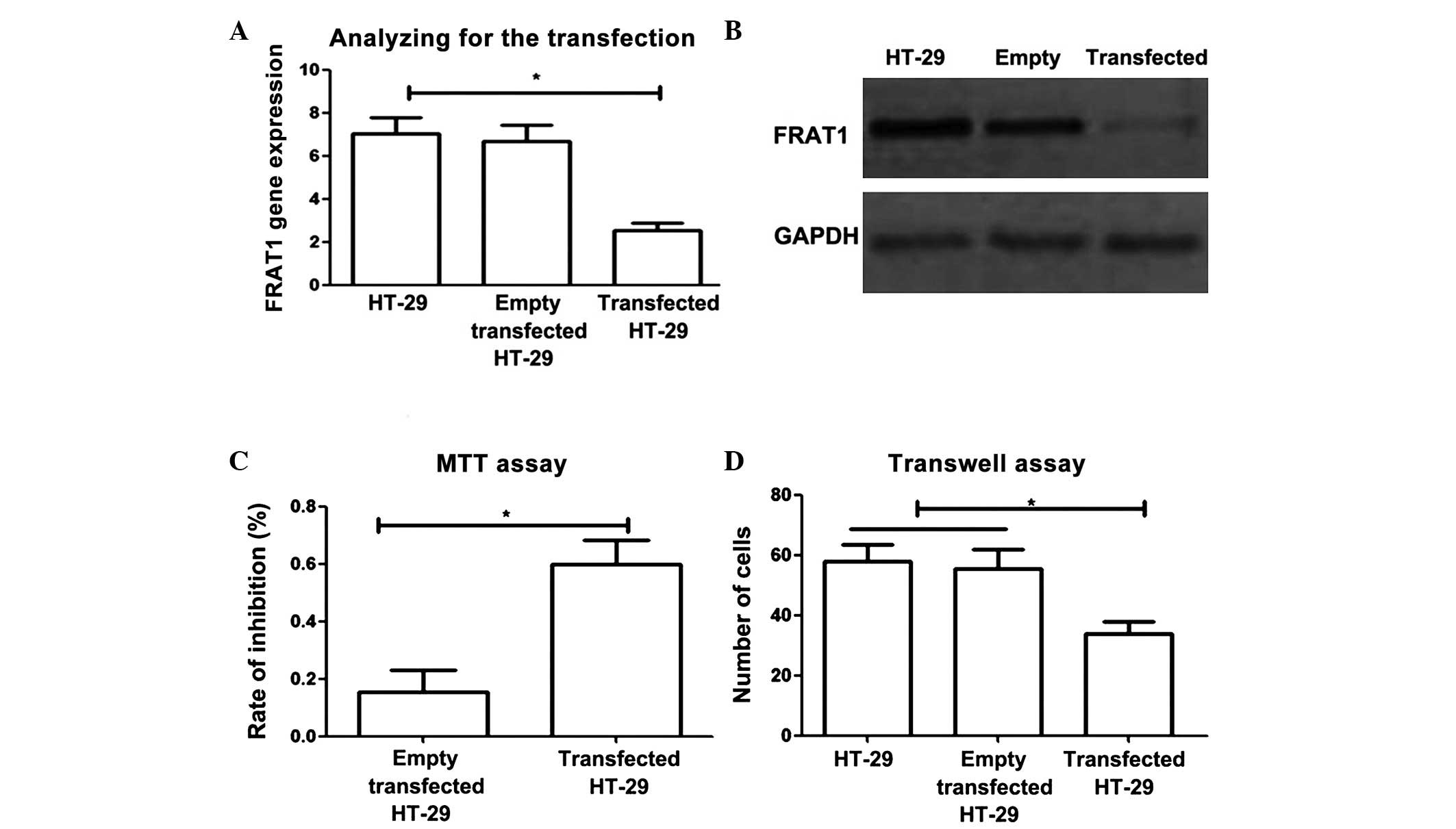
Small hairpin RNA (shRNA)-transfected HT-29 cells
were analyzed using PCR and western blotting to determine the
effects of reducing FRAT1 expression. The results revealed that
levels of FRAT1 gene and protein expression were significantly
decreased in transfected HT-29 cells (due to conversion of shRNA to
small interfering RNA) compared with control cells (P<0.05).
To investigate the effects of FRAT1 gene regulation
on the proliferation and migration of colon cancer cells, MTT and
Transwell assays were performed. MTT assay revealed that reducing
FRAT1 expression significantly inhibited the proliferation of colon
cancer cells (Fig. 2C; P<0.05).
Furthermore, Transwell assay demonstrated that in transfected HT-29
cells, the number of cells which passed through Matrigel within 12
h was significantly lower than that in normal HT-29 cells and empty
vector transfected HT-29 cells (P<0.05). These results indicated
that reducing FRAT1 expression significantly decreased the
migration ability of tumor cells (Fig.
2D).
Discussion
FRAT1 was originally identified as a T-cell lymphoma
proto-oncogene in mice (25). Recent
studies have confirmed that the abnormal activation of FRAT1
increased by competing with Axin for glycogen synthase kinase 3
phosphorylation sites binding, subsequently inhibiting the
degradation of Wnt/β-catenin phosphorylation and improving the
cytoplasmic secretion and retention of β-catenin. Subsequently, the
activation of β-catenin led to altered cell proliferation and
apoptosis (26–28).
High FRAT1 expression has been demonstrated in a
variety of tumors, including non-small cell lung cancer, ovarian
cancer, brain glioblastoma and other tumor tissues (14,29–31). High
FRAT1 expression is associated with the increased malignancy and a
higher clinical stage, thus FRAT1 affects tumor biological
characteristics (19). However,
studies investigating FRAT1 expression in digestive tract tissue
are limited. Previous studies have demonstrated an association
between increased FRAT1 expression and a higher TNM stage and
pathological stage in gastric cancer. However, further study is
required (16,18).
Previous studies using surgical specimens and
statistical analysis have been performed (1,27,28,32). In
the present study, gene and protein expression of FRAT1 was
analyzed in colon cancer specimens of different TNM and
pathological stages. The results showed that FRAT1 expression was
significantly higher in stage III and IV patients than stage I and
II patients (P<0.05). Furthermore, the pathological findings
revealed that FRAT1 expression was significantly increased in
poorly differentiated colon cancer tissues compared with that in
well- and moderately-differentiated colon cancer tissues
(P<0.05). Previous studies have also shown that in colon cancer,
FRAT1 expression is positively correlated with clinical and
pathological progression of tumors (33–35).
Taking into consideration a previous study, which investigated the
mechanism of colon cancer development (34), we hypothesize that in colon cancer,
regulation of the FRAT1-Wnt/β-catenin pathway occurs. Therefore,
similar to the non-small cell lung cancer, FRAT1 may present a
novel therapeutic target for colon cancer.
Although statistical analysis has revealed a
positive correlation between tumor progression and FRAT1
expression, such analysis does not show the direct effect of FRAT1
expression on tumor cells (36).
Therefore, in vitro shRNA interference experiments were
conducted to investigate the effects of FRAT1 expression on tumor
cells. An interference expression vector was constructed and
transfected into the human colon cancer cell line, HT-29, which
inhibited FRAT1 expression, as confirmed by RT-PCR and western blot
analysis. MTT and Transwell assays revealed that migration and
proliferation of the transfected colon cancer cells were
significantly decreased. Thus, regulation of FRAT1 expression may
be used to inhibit cancer cell proliferation and migration, which
are known to be associated with the clinicopathological stage.
Thus, we hypothesize that FRAT1 exerts an important role in the
development of colon cancer, and the inhibition of FRAT1 expression
effectively inhibits cancer cell progression.
In conclusion, the results of the present study
indicate that in colon cancer, FRAT1 gene and protein expression is
positively correlated with TNM and pathological stage. In addition,
a positive correlation was also identified between FRAT1 expression
and the degree of tumor malignancy. Further cell transfection
experiments revealed that inhibition of FRAT1 expression
significantly reduced proliferation and migration in colon cancer
cells. Therefore, FRAT1 may present an important tool for
evaluation and an important therapeutic target for the treatment of
colon cancer, however, further study is required.
References
|
1
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, D Naishadham D and Jemal A:
Cancer statistics, 2013. CA Cancer J Clin. 63:11–30. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bray F, Ren JS, Masuyer E and Ferlay J:
Global estimates of cancer prevalence for 27 sites in the adult
population in 2008. Int J Cancer. 132:1133–1145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rabeneck L, Horton S, Zauber AG and Earle
C: Colorectal cancerCancer: Disease Control Priorities. 3. 3rd.
Gelband H, Jha P, Sankaranarayanan R and Horton S: The World Bank;
Washington, D.C.: 2015
|
|
7
|
Zhang Y, Han Y, Zheng R, Yu JH, Miao Y,
Wang L and Wang EH: Expression of Frat1 correlates with expression
of β-catenin and is associated with a poor clinical outcome in
human SCC and AC. Tumor Biol. 33:1437–1444. 2012. View Article : Google Scholar
|
|
8
|
Tsikitis VL, Larson DW, Heubner M, Lohse
CM and Thompson PA: Predictors of recurrence free survival for
patients with stage II and III colon cancer. BMC Cancer.
14:3362014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hutchins G, Southward K, Handley K, Magill
L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M,
Kerr D, et al: Value of mismatch repair, KRAS, and BRAF mutations
in predicting recurrence and benefits from chemotherapy in
colorectal cancer. J Clin Oncol. 29:1261–1270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo G, Liu B, Zhong C, Zhang X, Mao X,
Wang P, Jiang X, Huo J, Jin J, Liu X and Chen X: FRAT1 expression
and its correlation with pathologic grade, proliferation, and
apoptosis in human astrocytomas. Med Oncol. 28:1–6. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong, GZ, Shim AR, Hyeon JS, Lee HJ and
Ryu JH: Inhibition of Wnt/β-catenin pathway by dehydrocostus
lactone and costunolide in colon cancer cells. Phytother Res.
29:680–686. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pai P, Rachagani S, Dhawan P and Batra SK:
Mucins and Wnt/β-catenin signaling in gastrointestinal cancers: An
unholy nexus. Carcinogenesis. 37:223–232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou FQ, Qi YM, Xu H, Wang QY, Gao XS and
Guo HG: Expression of EpCAM and Wnt/β-catenin in human colon
cancer. Genet Mol Res. 14:4485–4494. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo G, Mao X, Wang P, Liu B, Zhang X,
Jiang X, Zhong C, Huo J, Jin J and Zhuo Y: The expression profile
of FRAT1 in human gliomas. Brain Res. 1320:152–158. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bechard M, Trost R, Singh AM and Dalton S:
Frat is a phosphatidylinositol 3-kinase/Akt-regulated determinant
of glycogen synthase kinase 3β subcellular localization in
pluripotent cells. Mol Cell Biol. 32:288–296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goksel G, Bilir A, Uslu R, Akbulut H,
Guven U and Oktem G: WNT1 gene expression alters in heterogeneous
population of prostate cancer cells; decreased expression pattern
observed in CD133+/CD44+ prostate cancer stem cell spheroids. J
BUON. 19:207–214. 2014.PubMed/NCBI
|
|
17
|
Guo G, Kuai D, Cai S, Xue N, Liu Y, Hao J,
Fan Y, Jin J, Mao X, Liu B, et al: Knockdown of FRAT1 expression by
RNA interference inhibits human glioblastoma cell growth, migration
and invasion. PLoS One. 8:e612062013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Verheyen EM and Gottardi CJ: Regulation of
Wnt/β-catenin signaling by protein kinases. Dev Dyn. 239:34–44.
2010.PubMed/NCBI
|
|
19
|
Walf-Vorderwülbecke V, de Boer J, Horton
SJ, van Amerongen R, Proost N, Berns A and Williams O: Frat2
mediates the oncogenic activation of Rac by MLL fusions. Blood.
120:4819–4828. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang JX, Tong ZT, Yang L, Wang F, Chai
HP, Zhang F, Xie MR, Zhang AL, Wu LM, Hong H, et al: PITX2: A
promising predictive biomarker of patients' prognosis and
chemoradioresistance in esophageal squamous cell carcinoma. Int J
Cancer. 132:2567–2577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amar S, Belmaker RH and Agam G: The
possible involvement of glycogen synthase kinase-3 (GSK-3) in
diabetes, cancer and central nervous system diseases. Curr Pharm
Des. 17:2264–2277. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schröder C, Srinivasan H, Sill M,
Linseisen J, Fellenberg K, Becker N, Nieters A and Hoheisel JD:
Plasma protein analysis of patients with different B-cell lymphomas
using high-content antibody microarrays. Proteomics Clin Appl.
7:802–812. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vermeulen L, De Sousa E Melo F, van der
Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M,
Merz C, Rodermond H, et al: Wnt activity defines colon cancer stem
cells and is regulated by the microenvironment. Nat Cell Biol.
12:468–476. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jonkers J, Korswagen HC, Acton D, Breuer M
and Berns A: Activation of a novel proto-oncogene, Frat1,
contributes to progression of mouse T-cell lymphomas. EMBO J.
16:441–450. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu LW, Kawamoto EM, Brietzke E, Scavone C
and Lafer B: The role of Wnt signaling and its interaction with
diverse mechanisms of cellular apoptosis in the pathophysiology of
bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry.
35:11–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seira O and Del Río JA: Glycogen synthase
kinase 3 Beta (GSK3β) at the tip of neuronal development and
regeneration. Mol Neurobiol. 49:931–944. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Loke J, Pearlman A, Radi O, Zuffardi O,
Giussani U, Pallotta R, Camerino G and Ostrer H: Mutations in
MAP3K1 tilt the balance from SOX9/FGF9 to WNT/β-catenin signaling.
Hum Mol Genet. 23:1073–1083. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Yu JH, Lin XY, Miao Y, Han Y, Fan
CF, Dong XJ, Dai SD and Wang EH: Overexpression of Frat1 correlates
with malignant phenotype and advanced stage in human non-small cell
lung cancer. Virchows Arch. 459:255–263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Y, Hewitt SM, Liu S, Zhou X, Zhu H,
Zhou C, Zhang G, Quan L, Bai J and Xu N: Tissue microarray analysis
of human FRAT1 expression and its correlation with the subcellular
localisation of beta-catenin in ovarian tumours. Br J Cancer.
94:686–691. 2006.PubMed/NCBI
|
|
31
|
He L, Yang Z, Zhou J and Wang W: The
clinical pathological significance of FRAT1 and ROR2 expression in
cartilage tumors. Clin Transl Oncol. 17:438–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ornostay A, Cowie AM, Hindle M, Baker CJ
and Martyniuk CJ: Classifying chemical mode of action using gene
networks and machine learning: A case study with the herbicide
linuron. Comp Biochem Physiol Part D Genomics Proteomics.
8:263–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jensen K, Günther J, Talbot R, Petzl W,
Zerbe H, Schuberth HJ, Seyfert HM and Glass EJ: Escherichia
coli-and Staphylococcus aureus-induced mastitis differentially
modulate transcriptional responses in neighbouring uninfected
bovine mammary gland quarters. BMC Genomics. 14:362013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Domínguez F, Garrido T and Simón C:
Proteomics analysis of the endometrium and embryo. Can we improve
IVF outcome?Human Assisted Reproductive Technology: Future Trends
in Laboratory and Clinical Practice. Gardner DK, Rizk B and Falcone
T: Cambridge University Press; Cambridge, UK: pp. 2892011,
View Article : Google Scholar
|
|
35
|
Cayeux E: Safe mud pump management while
conditioning mud: On the adverse effects of complex heat transfer
and barite sag when establishing circulationProceedings of the IFAC
Workshop on Automatic Control in Offshore Oil and Gas Production.
Norwegian University of Science and Technology; Trondheim, Norway:
pp. 231–238. 2012
|
|
36
|
McIntyre RE, van der Weyden L and Adams
DJ: Cancer gene discovery in the mouse. Curr Opin Genet Dev.
22:14–20. 2012. View Article : Google Scholar : PubMed/NCBI
|