Introduction
Colorectal carcinoma (CRC) is a common clinical
malignancy, with an incidence that ranks as the second most
frequent malignant neoplasm in Western countries, and the incidence
of colorectal cancer in China is also rising, becoming the most
common malignancy (1). At present,
the primary treatment for colorectal cancer is surgery, and early
diagnosis and timely surgical treatment markedly improve the
survival rate of patients. Therefore, identifying key factors
involved in the development of cancer is necessary in order to not
only aid the diagnosis of cancer, but also to act as a prognostic
indicator in cancer patients.
Non-coding RNAs (ncRNAs) are found throughout the
genome, although the function of ncRNAs is only partially
understood. Numerous studies indicate that long ncRNAs (lncRNAs),
which are >200 nt in length, and microRNAs (miRNAs or miRs),
which are ~22 nt in length, have various functions in the
development and progression of cancer (2–8).
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is
an abundant nucleus-restricted lncRNA, and previous studies have
demonstrated that MALAT1 is upregulated in several solid tumors,
and associated with cancer metastasis and recurrence (9–14). All
these studies have revealed that MALAT1 plays an important role in
promoting tumorigenesis, and tumors expressing a high level of
MALAT1 appear to be more invasive and result in a poorer
prognosis.
miRNAs have been reported to be involved in
tumorigenesis (15,16), acting as tumor suppressors, such as
miRNA-15a, oncogenes, such as miRNA-21, or as promoters and
suppressors of metastasis, such as miRNA-182 and miRNA-126,
respectively. Other studies have described the altered expression
of miRNAs in cancer tissues compared with normal tissues (17,18),
suggesting that these miRNAs may potentially indicate novel
clinical diagnostic and prognostic markers. miRNA-619-5p
(miR-619-5p) is a miRNA that binds with high affinity to the
messenger (m)RNA of 1,215 genes, and miR-619-5p has binding sites
in the coding sequences and untranslated regions (UTRs) of mRNAs
(19).
Bioinformatics analysis indicated that miR-619-5p
has several binding sites on the 3′-UTR of MALAT1. However, the
association between MALAT1 and miR-619-5p in CRC is not well
studied. In the present study, the correlations between the
expression of MALAT1 and miR-619-5p was investigated, in addition
to the association between the clinicopathological features and
survival outcomes of patients with stage II and III CRC.
Materials and methods
Clinical samples
Paraffin-embedded tumors and adjacent normal tissue
samples from 120 patients with CRC who underwent tumor resection at
Nantong Second People's Hospital (Nantong, Jiangsu, China), Nantong
Tumor Hospital (Nantong, Jiangsu, China) or Affiliated Hospital of
Nantong University (Nantong, Jiangsu, China) between 2006 and 2010
were collected. All patients had not received chemotherapy or
radiotherapy prior to surgery. Each tissue sample was obtained
under sterile conditions by removing the CRC tissue and normal
colon mucosa along the surgical margin. The present study was
approved by the ethics committees of Nantong Second People's
Hospital, Nantong Tumor Hospital, and Affiliated Hospital of
Nantong University. All patients provided written informed
consent.
Total RNA and miRNA isolation
Total RNA was extracted from tissues using TRIzol
reagent (Takara Biotechnology Co., Ltd., Dalian, China) and
complementary DNA was synthesized using PrimeScript™ RT Reagent kit
with gDNA Eraser (Perfect Real Time) obtained from Takara
Biotechnology Co., Ltd., according to the manufacturer's protocol.
miRNA was extracted from the tissue using the Ambion mirVana™ miRNA
Isolation kit (catalog no., AM1560; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). All preparation and handling steps for RNA
isolation were performed under RNase-free conditions.
Quantitative polymerase chain reaction
(qPCR)
All primers were designed and synthesized by
Shanghai Sangon Biological Engineering Technology & Services
Co., Ltd. (Shanghai, China), and the primer sequences were as
follows: MALAT1 forward, 5′-AGGCGTTGTGCGTAGAGGA-3′ and reverse,
5′-GGATTTTTACCAACCACTCGC-3′; glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) forward, 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and
reverse, 5′-CCAAATTCGTTGTCATACCAGGAAATG-3′; miR-619-5p forward,
5′-GCUGGGAUUACAGGCAUGAGCC-3′ and reverse,
5′-TCTACGTCGTATCGTCATCTGAC-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
SYBR® Premix Ex Taq™ (Tli RNaseH Plus), ROX plus from
Takara Biotechnology Co., Ltd. was used according to the
manufacturer's protocol, and the total reaction volume was 20 µl,
which contained 10 µl SYBR Green premix, 0.3 µl correction dye, 1.5
µl cDNA, 0.5 µl forward primer and 0.5 µl reverse primer, and water
was added to make a final volume of 20 µl. The reaction protocol
was as follows: 95°C for 5 min; 40 cycles at 95°C for 15 sec; 60°C
for 15 sec; and 72°C for 15 sec. qPCR was performed using the
Applied Biosystems 7300 Real-Time PCR System (Thermo Fisher
Scientific, Inc.). The 2−ΔΔCq method (20) was used for quantification of the PCR
results All assays were performed in triplicate and independently
repeated three times.
Statistical analysis
Statistical analysis was performed using SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA) and the values were
expressed as the mean ± standard deviation. Comparisons of
continuous data between the two groups were performed using an
independent t-test, and categorical data were analyzed using
the χ2 test or Fisher's exact test. The overall survival
(OS) time was calculated as the time between the date of surgery
and the date of mortality, or the last follow-up. The disease-free
survival (DFS) time was defined as the time between the date of
surgery and the date of local or distant recurrence or the date of
the last follow-up. Patient survival rates were calculated using
the Kaplan-Meier method, and statistically significant differences
in survival were identified using the log-rank test. Correlations
between the genes were analyzed using Spearman's rank correlation
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
MALAT1 expression in CRC tissues and
adjacent normal tissues
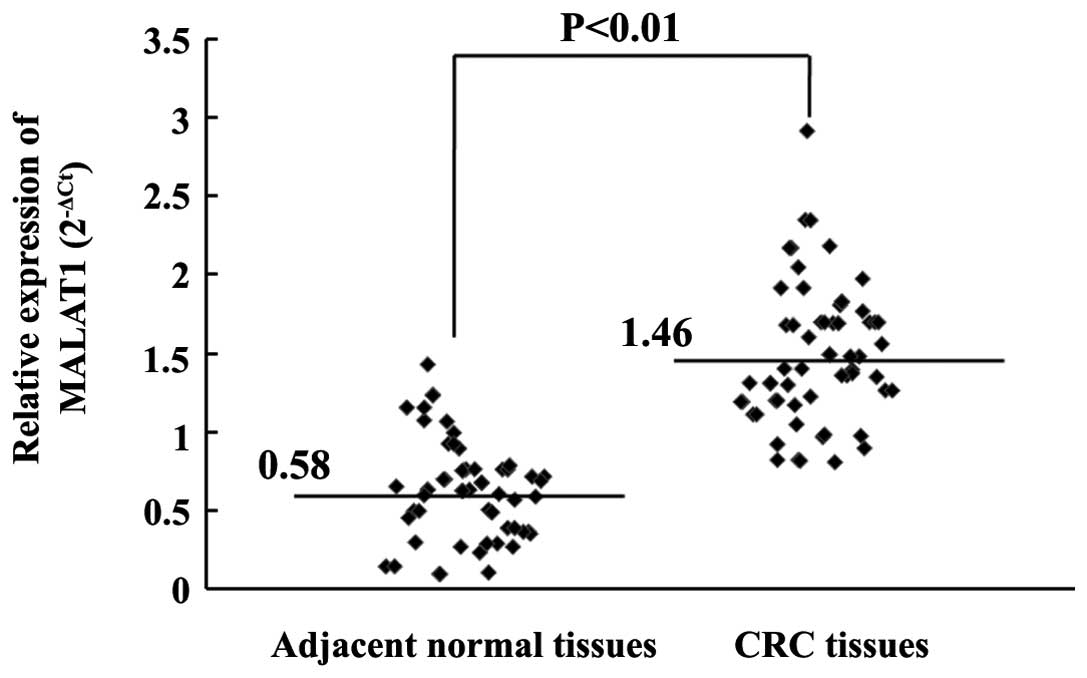
The expression levels of MALAT1 in 120 CRC tissues
and adjacent normal tissues were examined by reverse transcription
(RT)-qPCR. The levels of MALAT1 in the CRC tissues were 2.52 times
higher compared with the levels in the adjacent normal tissues,
which was a significant difference (P<0.01; Fig. 1). These results demonstrated that
MALAT1 was evidently upregulated in CRC tumors.
miR-619-5p expression in CRC tissues
and adjacent normal tissues
The expression levels of miR-619-5p in 120 CRC
tissues and adjacent normal tissues were also determined by
RT-qPCR. The levels of miR-619-5p in CRC tissues were 5.79 times
lower compared with the levels measured in the adjacent normal
tissues, which was a significant difference (P<0.01; Fig. 2). The results demonstrated that
miR-619-5p was markedly downregulated in CRC tumors.
Association between MALAT1 expression
and the clinicopathological features of CRC patients
Subsequently, the expression of MALAT1 was examined
in 120 patients with stage II and III CRC. According to the
expression of MALAT1, these cases were divided into the high MALAT1
expression group (n=60) and low expression group (n=60), based on
the MALAT1/GAPDH ratio in CRC tissues. The expression of MALAT1 was
significantly increased in male patients compared with female
patients (P=0.027), and MALAT1 expression was significantly
associated with the tumor-node-metastasis (TNM) stage (P=0.019),
lymphovascular invasion (P=0.047) and perineural invasion
(P=0.012), although no association was found between MALAT1
expression and the other clinicopathological features (Table I).
 | Table I.Association between the expression
level of MALAT1 and the clinicopathological features of the 120
patients with colorectal cancer. |
Table I.
Association between the expression
level of MALAT1 and the clinicopathological features of the 120
patients with colorectal cancer.
| Clinical
features | High MALAT1
expression, n (%) | Low MALAT1
expression, n (%) | P-value |
|---|
| Total | 60
(100.00) | 60
(100.00) |
|
| Gender |
|
| 0.027 |
| Male | 41 (68.33) | 28 (46.67) |
|
|
Female | 19 (31.67) | 32 (53.33) |
|
| Age |
|
| 0.473 |
| <65
years | 37 (61.67) | 39 (65.00) |
|
| ≥65
years | 23 (38.33) | 21 (35.00) |
|
| Tumor size,
diameter |
|
| 0.072 |
| ≤6
cm | 22 (36.67) | 29 (48.33) |
|
| >6
cm | 38 (63.33) | 31 (51.67) |
|
| Tumor site |
|
| 0.746 |
|
Rectum | 39 (65.00) | 41 (68.33) |
|
|
Colon | 21 (35.00) | 19 (31.67) |
|
| Tumor histology |
|
| 0.344 |
|
Adenocarcinoma | 43 (71.67) | 38 (63.33) |
|
| Mucinous
adenocarcinoma | 17 (28.33) | 22 (36.67) |
|
| Tumor
differentiation |
|
| 0.239 |
|
Well/moderate | 31 (60.00) | 36 (51.67) |
|
| Poor | 29 (40.00) | 24 (48.33) |
|
| TNM stage |
|
| 0.019 |
| Stage
II | 18 (30.00) | 28 (46.67) |
|
| Stage
III | 42 (70.00) | 32 (53.33) |
|
| Lymph vascular
invasion |
|
| 0.047 |
|
Absence | 41 (68.33) | 31 (51.67) |
|
|
Presence | 19 (31.67) | 29 (48.33) |
|
| Perineural
invasion |
|
| 0.011 |
|
Absence | 46 (76.67) | 33 (55.00) |
|
|
Presence | 14 (23.33) | 27 (45.00) |
|
Association between miR-619-5p
expression and clinicopathological features of CRC patients
The association between miR-619-5p expression and
the clinicopathological features of CRC patients was also
investigated. The expression of miR-619-5p was significantly
decreased in male patients compared with female patients (P=0.032),
and the expression of miR-619-5p was significantly associated with
TNM stage (P=0.012), lymphovascular invasion (P=0.023) and
perineural invasion (P=0.009), while no association was identified
between miR-619-5p expression and other clinicopathological
features (Table II).
 | Table II.Association between miR-619-5p
expression and clinicopathological features of the 120 patients
with colorectal cancer. |
Table II.
Association between miR-619-5p
expression and clinicopathological features of the 120 patients
with colorectal cancer.
| Clinical
features | High miR-619-5p
expression, n (%) | Low miR-619-5p
expression, n (%) | P-value |
|---|
| Total | 60
(100.00) | 60
(100.00) |
|
| Gender |
|
| 0.032 |
|
Male | 29 (48.33) | 40 (66.67) |
|
|
Female | 31 (51.67) | 20 (33.33) |
|
| Age |
|
| 0.373 |
| <65
years | 40 (66.67) | 36 (60.00) |
|
| ≥65
years | 20 (33.33) | 24 (40.00) |
|
| Tumor size,
diameter |
|
| 0.057 |
| ≤6
cm | 30 (50.00) | 21 (35.00) |
|
| >6
cm | 30 (50.00) | 39 (65.00) |
|
| Tumor site |
|
| 0.328 |
|
Rectum | 42 (70.00) | 38 (63.33) |
|
|
Colon | 18 (30.00) | 22 (36.67) |
|
| Tumor
histology |
|
| 0.312 |
|
Adenocarcinoma | 39 (65.00) | 42 (70.00) |
|
|
Mucinous adenocarcinoma | 21 (35.00) | 18 (30.00) |
|
| Tumor
differentiation |
|
| 0.081 |
|
Well/moderate | 37 (61.67) | 30 (50.00) |
|
|
Poor | 23 (38.33) | 30 (50.00) |
|
| TNM stage |
|
| 0.012 |
| Stage
II | 27 (45.00) | 16 (26.67) |
|
| Stage
III | 33 (55.00) | 44 (73.33) |
|
| Lymph vascular
invasion |
|
| 0.023 |
|
Absence | 29 (48.33) | 39 (65.00) |
|
|
Presence | 31 (51.67) | 21 (35.00) |
|
| Perineural
invasion |
|
| 0.009 |
|
Absence | 31 (51.67) | 45 (75.00) |
|
|
Presence | 29 (48.33) | 15 (25.00) |
|
Univariate analysis of prognostic
factors in patients with stage II and III CRC
The median follow-up period for the 120 patients
with CRC in the present study was 53.6 months, with a range of
10–76.4 months. MALAT1 expression, miR-619-5p expression, TNM stage
and perineural invasion were significantly associated with the DFS
and OS times (Table III). In
particular, patients with a high level of MALAT1 expression or low
level of miR-619-5p expression possessed a significantly shorter
DFS (P=0.002) and OS (P=0.004) time compared with patients with low
MALAT1 expression or a high level of miR-619-5p expression, and
patients with perineural invasion demonstrated significantly
shorter DFS (P=0.001) and OS (P=0.003) times compared with patients
without perineural invasion. Additionally, patients in with TNM
stage III CRC also experienced a significantly shorter OS time
(P=0.037) compared with patients with TNM stage II CRC.
 | Table III.Univariate analysis of DFS and OS for
the 120 patients with colorectal cancer. |
Table III.
Univariate analysis of DFS and OS for
the 120 patients with colorectal cancer.
| Clinical
features | DFS, P-value | OS, P-value |
|---|
| MALAT1
expression |
|
|
|
High | 0.002 | 0.004 |
|
Low |
|
|
| miR-619-5p
expression |
|
|
|
High | 0.002 | 0.004 |
|
Low |
|
|
| Gender |
|
|
|
Male | 0.898 | 0.914 |
|
Female |
|
|
| Age |
|
|
| <65
years | 0.372 | 0.249 |
| ≥65
years |
|
|
| Tumor size,
diameter |
|
|
| ≤6
cm | 0.141 | 0.095 |
| >6
cm |
|
|
| Tumor site |
|
|
|
Rectum | 0.484 | 0.596 |
|
Colon |
|
|
| Tumor
histology |
|
|
|
Adenocarcinoma | 0.842 | 0.529 |
|
Mucinous adenocarcinoma |
|
|
| Tumor
differentiation |
|
|
|
Well/moderate | 0.565 | 0.683 |
|
Poor |
|
|
| TNM stage |
|
|
| Stage
II | 0.152 | 0.037 |
| Stage
III |
|
|
| Lymph vascular
invasion |
|
|
|
Absence | 0.214 | 0.075 |
|
Presence |
|
|
| Perineural
invasion |
|
|
|
Absence | 0.001 | 0.003 |
|
Presence |
|
|
Correlation analysis of the MALAT1 and
miR-619-5p expression in CRC tissues
Since the upregulated MALAT1 expression and
downregulated miR-619-5p expression in CRC tissues were each
associated with gender, TNM stage, lymphovascular invasion and
perineural invasion, it is essential to investigate the correlation
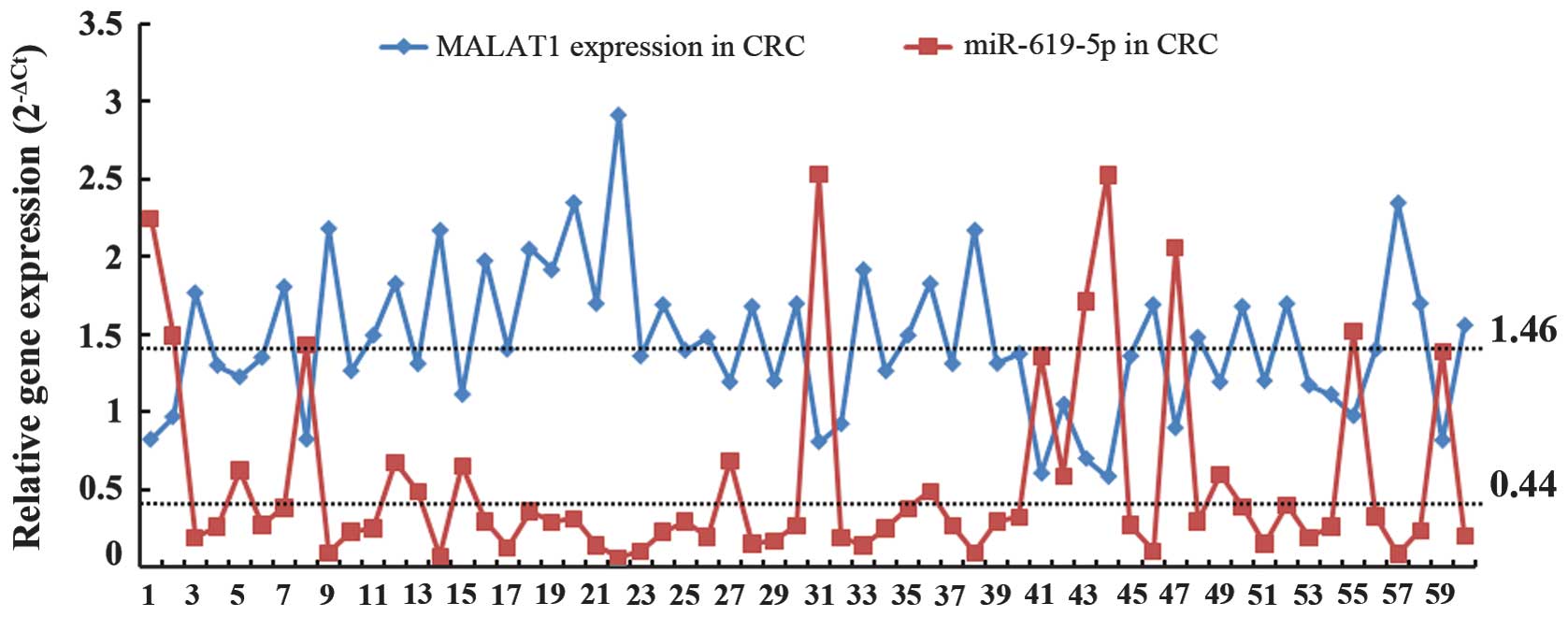
between MALAT1 expression and miR-619-5p expression. As
demonstrated in Fig. 3 and Table IV, MALAT1 expression was found to be
negatively correlated with miR-619-5p expression (r=−0.415,
P=0.004) in CRC tissues. In general, if MALAT1 expression was high,
miR-619-5p expression was low in CRC tissues.
 | Table IV.Spearman's rank correlation analysis
of MALAT1 and miR-619-5p expression. |
Table IV.
Spearman's rank correlation analysis
of MALAT1 and miR-619-5p expression.
|
| Correlation |
|---|
|
|
|
|---|
| Parameters | r | P-value |
|---|
| MALAT1
expression | −0.415 | 0.004a |
| miR-619-5p
expression |
|
|
Discussion
CRC is one of the most common human malignant
cancers, which are the synergistic results of various oncogenes and
tumor suppressor genes, and remains the third leading cause of
cancer-associated death worldwide (1). Although recent advances have improved
the diagnosis and therapy of patients with CRC, there are few
reliable markers available to accurately predict metastasis in CRC
patients, particularly in patients with early-stage CRC.
Previously, lncRNAs have been increasingly reported
to be involved in human disease (21,22).
MALAT1 is a highly conserved and newly identified lncRNA, which was
first found to be overexpressed in metastatic non-small cell lung
cancer (NSCLC) (9). Subsequently,
MALAT1 was found to be associated with several other cancers,
including cervical, hepatic, breast and renal cancer (10,11,23–26),
and was considered to regulate tumor growth, invasion and migration
in different types of cancers (27–30).
In addition, short ncRNAs, such as miRNAs, have been
the focus of studies, and numerous miRNAs have shown extremely
important effects in the development of cancers. miR-619-5p is a
miRNA that binds with high affinity to the mRNAs of 1,215 genes,
and miR-619-5p has binding sites in the coding sequences and UTRs
of mRNAs (19). Bioinformatics
analysis indicated that miR-619-5p has several binding sites on the
3′-UTR of MALAT1. However, the association between MALAT1 and
miR-619-5p in CRC is not well studied.
The present study examined the expression of MALAT1
and miR-619-5p in CRC tissues and adjacent normal tissues. The
analysis indicated that there were significant differences between
MALAT1 expression in CRC tissues and adjacent normal tissues, which
were consistent with previously reported results (13,14,31), and
to the best of our knowledge, miR-619-5p expression was reported to
be significantly different between CRC tissues and adjacent normal
tissues for the first time in the present study.
Investigation of the molecular diagnosis of CRC may
broaden the scope of medical research. The present results have
demonstrated that patients with high MALAT1 expression and low
miR-619-5p expression had a significantly increased risk of
metastasis, such as lymphovascular and perineural invasion,
subsequent to radical surgery. MALAT1 was first found to be
associated with tumor metastasis in patients with NSCLC, and
knockdown of MALAT1 in A549 lung cancer cells inhibits cell
migration without affecting cell proliferation (30). Additional mechanism analysis revealed
that MALAT1 is critical for the Wnt/β-catenin signaling pathway and
releasing the oncogene polypyrimidine tract binding protein-2
(PTBP-2) from the splicing factor proline/glutamine-rich/PTBP-2
complex (13,14). Overall, these mechanisms may explain
the strong tendency for metastasis subsequent to surgery in
patients with high expression of MALAT1. In addition, miR-619-5p
has been found to be extremely important in the metastasis of CRC,
whose function and mechanism required additional investigation.
Since miR-619-5p has several binding sites on the 3′-UTR of MALAT1,
assessment of the function and mechanism of miR-619-5p may be
extremely beneficial in future studies.
Univariate Cox regression analysis indicated that
MALAT1 expression, miR-619-5p expression and perineural invasion
were independent predictors of the DFS and OS times, and tumor TNM
stage and lymphovascular invasion were also found to be independent
predictors of the OS time.
Since upregulated MALAT1 expression and
downregulated miR-619-5p expression in CRC tissues were each
associated with the TNM stage, metastasis, DFS time and OS time,
the correlation between MALAT1 expression and miR-619-5p expression
was investigated. Spearman's rank correlation analysis revealed a
negative correlation between the differential expression of MALAT1
and miR-619-5p (r=−0.346; P=0.030), suggesting that combined
detection of miR-619-5p and MALAT1 may improve the accuracy of the
diagnosis of CRC, and the expression of miR-619-5p and MALAT1 may
act as a good prognostic indicator in CRC patients.
In conclusion, upregulated MALAT1 expression and
downregulated miR-619-5p expression may be involved in the
progression of CRC and may therefore be considered a diagnostic
marker and prognostic factor for patients with stage II or III CRC.
The identification of this novel biomarker may aid the
understanding of the possible molecular mechanisms underlying the
recurrence and metastasis of CRC, and provide additional
therapeutic targets for CRC patients.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pasmant E, Sabbagh A, Masliah-Planchon J,
Ortonne N, Laurendeau I, Melin L, Ferkal S, Hernandez L, Leroy K,
Valeyrie-Allanore L, et al: Role of noncoding RNA ANRIL in genesis
of plexiform neurofibromas in neurofibromatosis type 1. J Natl
Cancer Inst. 103:1713–1722. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamada K, Kano J, Tsunoda H, Yoshikawa H,
Okubo C, Ishiyama T and Noguchi M: Phenotypic characterization of
endometrial stromal sarcoma of the uterus. Cancer Sci. 97:106–112.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tseng JJ, Hsieh YT, Hsu SL and Chou MM:
Metastasis associated lung adenocarcinoma transcript 1 is
up-regulated in placenta previa increta/percreta and strongly
associated with trophoblast-like cell invasion in vitro. Mol Hum
Report. 15:725–731. 2009. View Article : Google Scholar
|
|
13
|
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L,
Sun J, Cai J, Qin J, Ren J and Li Q: Resveratrol Inhibits invasion
and metastasis of colorectal cancer cells via MALAT1 mediated
Wnt/β-catenin signal pathway. PLoS One. 8:e787002013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT-1
promotes tumor growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP-2 from SFPQ/PTBP-2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Skaftnesmo KO, Prestegarden L, Micklem DR
and Lorens JB: MicroRNAs in tumorigenesis. Curr Pharm Biotechnol.
8:320–325. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hernando E: microRNAs and cancer: Role in
tumorigenesis, patient classification and therapy. Clin Transl
Oncol. 9:155–160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Faragalla H, Youssef YM, Scorilas A,
Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A,
Lichner Z, et al: The clinical utility of miR-21 as a diagnostic
and prognostic marker for renal cell carcinoma. J Mol Diagn.
14:385–392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koberle V, Kronenberger B, Pleli T, Trojan
J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem
S, Piiper A and Waidmann O: Serum microRNA-1 and microRNA-122 are
prognostic markers in patients with hepatocellular carcinoma. Eur J
Cancer. 49:3442–3449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ivashchenko A, Berillo O, Pyrkova A,
Niyazova R and Atambayeva S: The properties of binding sites of
miR-619-5p, miR-5095, miR-5096 and miR-5585-3p in the mRNAs of
human genes. Biomed Res Int. 2014:7207152014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Esteller M: Non-coding RNAs in human
disease. Nature Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luo JH, Ren B, Keryanov S, Tseng GC, Rao
UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, et al:
Transcriptomic and genomic analysis of human hepatocellular
carcinomas and hepatoblastomas. Hepatology. 44:1012–1024. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guffanti A, Iacono M, Pelucchi P, Kim N,
Soldà G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ,
et al: A transcriptional sketch of a primary human breast cancer by
454 deep sequencing. BMC Genomics. 10:1632009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Praz V, Jagannathan V and Bucher P:
CleanEx: A database of heterogeneous gene expression data based on
a consistent gene nomenclature. Nucleic Acids Res. 32:(Database
issue). D542–D547. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davis IJ, Hsi BL, Arroyo JD, Vargas SO,
Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi
M, et al: Cloning of an Alpha-TFEB fusion in renal tumors harboring
the t(6;11) (p21;q13) chromosome translocation. Proc Natl Acad Sci
USA. 100:6051–6056. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, Hillejan L, et al: The long noncoding MALAT-1 RNA
indicates a poor prognosis in non-small cell lung cancer and
induces migration and tumor growth. J Thorac Oncol. 6:1984–1892.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol
Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|