Introduction
Human osteosarcoma is the most common primary bone
malignancy sarcoma found primarily in children and young people
(1–3).
The patients with osteosarcoma exhibit severe clinical
manifestations, significantly diminished mobility and a reduced
life expectancy (4,5). However, the prognosis remained poor. It
is important to clarify the underlying mechanism so that safe and
effective therapeutic strategies may be developed.
MicroRNA (miRNA) is a class of small non-coding RNA,
with lengths of 18–22 nucleotides; they play important roles in
numerous biological processes through transcriptional suppression
of target genes (6–8). The critical functions of numerous miRNAs
in cancer biology have been illustrated, including the regulation
of various cellular processes, such as proliferation and apoptosis
(3,5,9,10). The abnormality of miR-574-3p was
observed in patients with non-small cell lung cancer and prostate
cancer, and it was found that this miRNA regulated colorectal
cancer growth and plays important roles in bladder cancer cell
lines (11–13). Nevertheless, whether miR-574-3p
participates in the progression of osteosarcoma remains unclear. In
the present study, it was demonstrated that the level of miR-574-3p
was significantly higher in the human osteosarcoma tissues and cell
lines and miR-574-3p also regulated osteosarcoma cell growth
through targeting the mothers against decapentaplegic homolog 4
(SMAD4) signaling pathway.
Materials and methods
Patients
Surgical specimens from 10 osteosarcoma and matched
normal control tissues were obtained postoperatively in May 2010
from the Department of Orthopedics of Jinling Hospital, Nanjing
University, School of Medicine (Nanjing, China) (3,5,10). All patients provided signed informed
consent for their tissues to be used for scientific research.
Ethical approval for the study was obtained from Jinling Hospital,
Nanjing University. All diagnoses were based on pathological and/or
cytological evidence (10). The
histological features of the specimens were evaluated by senior
pathologists according to the World Health Organization
classification criteria (11–13). Tissues were obtained prior to
chemotherapy and radiotherapy, and were immediately frozen and
stored at −80°C prior to reverse-transcription quantitative
polymerase chain reaction (RT-qPCR) assay (3,5,10).
Cell lines, cell culture and
transfection
Human osteosarcoma U2OS, SAOS and MG63 cell lines,
and the normal human osteoblast cell line were obtained from
Jinling Hospital Affiliated to Nanjing University (Nanjing, China),
and maintained in Dulbecco's Modified Eagle's medium supplemented
with 10% fetal bovine serum (GE Healthcare Bio-Sciences-
Pittsburgh, PA, USA) (10). U2OS and
MG63 cells were seeded in 24-well plates at a density of
6×104 cells per well and transfected with antisense
(ASO)-miR574, mimics (Shanghai GenePharma Co., Ltd., Shanghai,
China) or pcDNA3.1-SMAD4 at a concentration of 50 nM using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.,
Pittsburgh, PA, USA) according to the manufacturer's protocol.
Subsequent to 48 h, cells were used for additional experiments
(3).
RNA extraction and RT-qPCR
RNA from tumor tissues of differently treated groups
were homogenized in TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc.) and isolated according to the manufacturer's protocol.
RT-qPCR was assessed using the miScript RT Kit and miScript PCR
system (both Qiagen China Co., Ltd., Shanghai, China) according to
the manufacturer's protocol, during which glyceraldehyde
3-phosphate dehydrogenase was used as an internal control (3). The thermocycling conditions for the PCR
were was as follows: 95°C for 15 min; 40 cycles of 94°C for 15 sec,
55°C for 30 sec and 70°C for 30 sec; and 95°C for 1 min (3–5). Data were
normalized using the 2−ΔΔCq method, whereby
the cycle threshold (Cq) for the gene of interest was compared with
the Cq of the internal control gene (14).
Western blot assay
Cells were homogenized in Mammalian Protein
Extraction reagent containing a Halt Protease Inhibitor Cocktail
(both Thermo Fisher Scientific, Inc.), 150 mM NaCl and 1 mM EDTA.
The lysates were mixed with an equal volume of sample buffer (cat.
no. P0015B; Beyotime Institute of Biotechnology, Haimen, China) and
proteins were subsequently denatured by boiling. Protein
quantification was performed using an Enhanced BCA Protein Assay
kit (cat. no. P0010S; Beyotime Institute of Biotechnology). A total
of 20 µg protein was loaded into each lane. Total protein was
separated using SDS-PAGE on a 10% gel (3). The proteins were transferred onto
nitrocellulose membranes (GE Healthcare Life Sciences, Chalfont,
UK), blocked with 5% milk and incubated overnight at 4°C with the
primary antibody anti-SMAD4 (1:1,500; cat. no. ab40759; Abcam,
Cambridge, UK); β-actin was used as the loading control
(anti-β-actin; 1:13,000; cat. no. ab8227; Abcam). The blots were
then incubated with the anti-rabbit immunoglobulin G horseradish
peroxidase-conjugated secondary antibody (1:10,000; cat. no.
ab7090; Abcam) for 1 h at room temperature. Immunoreactivity was
detected using the ImageQuant™ LAS-4000 Luminescent Image Analyzer
(GE Healthcare Life Sciences). Protein bands were quantified by
means of densitometry using Fuji Image Gauge software (version 4.0;
Fujifilm) as previously described (3).
Proliferation assay
Cells were seeded into 96-well plates at a density
of 4,000 cells per well and transfected with miR-574-3p/negative
control/pcDNA3.1-SMAD4 (3,5,10). Cell
viability was determined by methyl thiazolyl tetrazolium (MTT)
assay to detect the viable proliferating cells at different time
points subsequent to transfection (3). The MTT Cell Proliferation and
Cytotoxicity assay kit (Amresco, LLC, Solon, OH, USA) was used.
Following photosensitization, 100 µg MTT in 20 µl PBS was added to
each well at 0, 24, 48 and 72 h and plates were subsequently
incubated at 37°C for 3 h. The reaction was stopped by the addition
of 180 µl dimethyl sulfoxide. The optical density (OD) of each
sample was subsequently measured at a wavelength of 490 nm using a
microplate spectrophotometer (Thermo 3001; Thermo Fisher
Scientific, Inc.). Cell survival rate (%) was calculated as the
following: (ODtreated / ODcontrol) × 100.
Apoptosis assay
Following transfection of the U2OS and MG-63 cells
with the miR-574-3p ASO, miR-574-3p mimics or controls. Cell
apoptosis was determined using apoptosis detection kit (Beyotime
Institute of Biotechnology) according to the manufacturer's
protocol (3,5,10). Samples
were determined by flow cytometry assays and the results were
analyzed using Cell-Quest software (version 7.5.3; Becton
Dickinson, San Jose, CA, USA).
miRNA target prediction and luciferase
assay
miRNA targets were predicted using the TargetScan
algorithms (http://www.targetscan.org). The
SMAD4-luciferase-3′-untranslated region (UTR) reporter was
generated by inserting the full length SMAD4 3′-UTR into pGL3
promoter vector (Promega Corporation, Madison, WI, USA) as
previously reported (3). Luciferase
assays were performed using NIH3T3 cells, as previously described
(15). Cells were transfected with
appropriate plasmids [TK Renilla luciferase (RL) plasmids,
wild type or mutated SMAD4 3′-UTR pGL3 promoter vector, synthetic
miR mimics/inhibitors (ASO oligo) with scramble control] in 24-well
plates. Cells were lysed for the luciferase assay 48 h subsequent
to transfection. Luciferase assays were conducted by using a
luciferase assay kit (E1910; Promega Corporation) according to the
manufacturer's protocol (3,5,10).
Statistical analysis
The data was expressed as the mean ± standard
deviation. Student's t-test was used for comparisons between
experimental groups and relevant controls (3,5). The
difference between groups was analyzed using one-way analysis of
variance when ≥3 groups were compared. P<0.05 was considered to
indicate a statistically significant difference. The correlation
between SMAD4 protein and miR-574-3p expression levels was assessed
by Spearman's rank correlation coefficient analysis.
Results
miR-574 level was elevated in human
osteosarcoma tissues
Initially, the present study assessed the levels of
miR-574-3p in U2OS, SAOS2 and MG63 osteosarcoma cell lines by
RT-qPCR; the miR-574-3p level in osteoblast cells was used as a
control. It was found that the expression levels of miR-574-3p in
the 3 osteosarcoma cell lines were significantly higher than that
in osteoblast cells (P=0.0023; Fig.
1A). The miR-574-3p levels in 10 pairs of human osteosarcoma
tissues and the matched tumor adjacent normal tissues were assayed
by RT-qPCR. It was found that osteosarcoma tissues showed a
significantly higher level of miR-574-3p than the matched
tumor-adjacent normal tissues in all 10 pairs (P=0.0089; Fig. 1B).
Downregulation of miR-574-3p inhibited
cell growth and induced cell apoptosis
To investigate the effects of miR-574-3p on
osteosarcoma cells, miR-574-3p levels were inhibited in U2OS and
MG-63 cells by transfecting the cells with miR-574-3p ASO. The
miR-574-3p levels in cells were assessed by RT-qPCR 48 h subsequent
to transfection. It was found that miR-574-3p ASO significantly
inhibited the miR-574-3p levels in the two cell lines (P=0.0073;
Fig. 2A). Following transfection,
cellular proliferation was then assessed by MTT analysis. The data
suggested that knockdown of miR-574-3p significantly inhibited the
growth of U2OS and MG63 cells (P=0.0102; Fig. 2B). Fluorescence-activated cell sorting
(FACS) analysis showed that the inhibition of miR-574-3p induced
cell apoptosis in MG63 and U2OS cells (Fig. 2C).
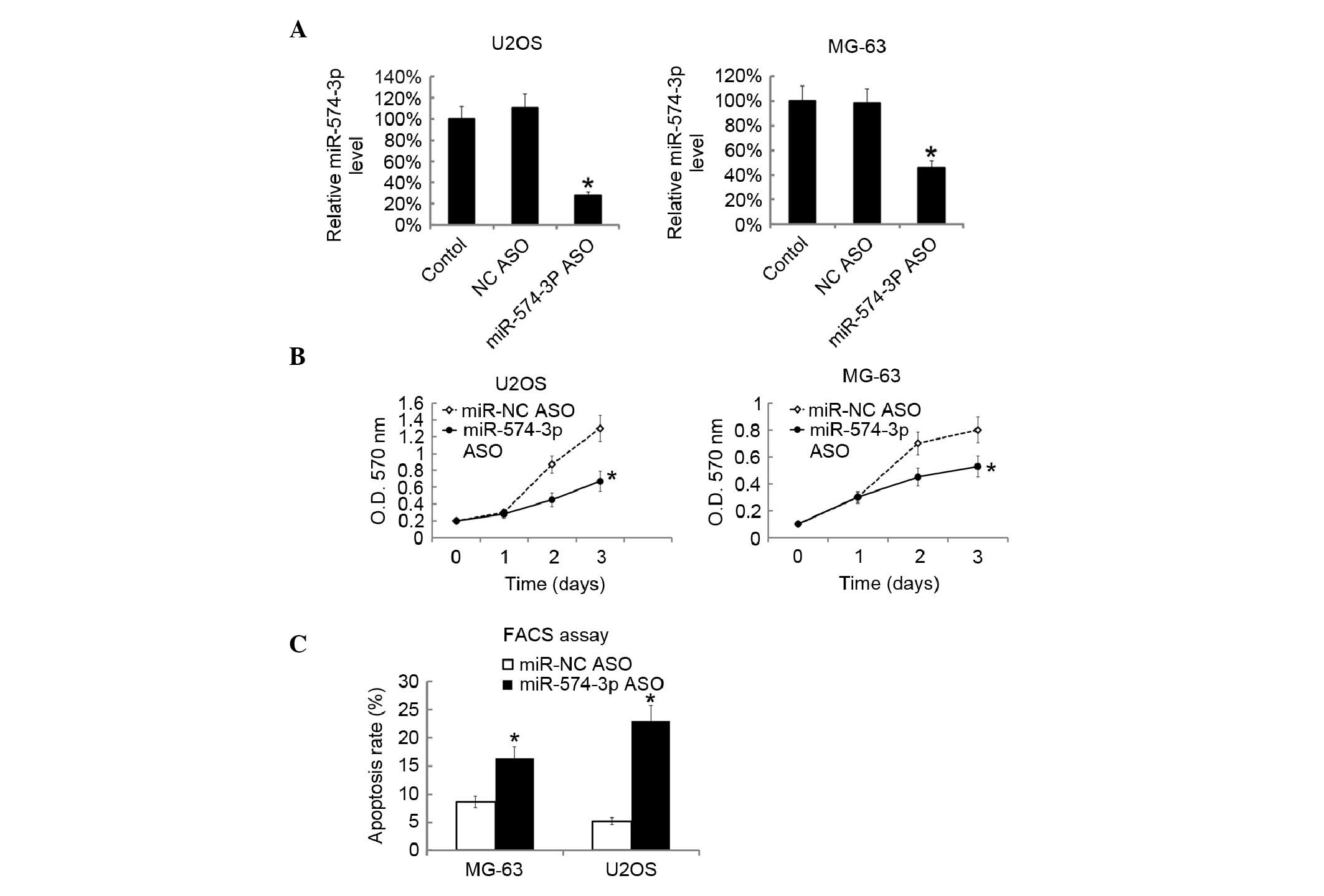 | Figure 2.Suppression of miR-574-3p inhibited
the cellular proliferation of and induced cells apoptosis of U2OS
and MG63 cells. U2OS and MG-63 cells were transfected with
miR-574-3p ASO. 48 h later miR-574-3p level in U2OS and MG-63 cells
was assayed by RT-qPCR. (A) The miR-574-3p level in the blank
control was arbitrarily defined as 100%. (B) Following miR-574-3p
ASO transfection, the proliferation of U2OS and MG-63 cells was
analyzed by MTT at the indicated time point. (C) Subsequent to 48 h
following miR-574-3p ASO transfection, apoptosis of U2OS and MG-63
cells was assessed by FACS analysis. All data are expressed as the
mean ± SD, with experiments repeated three times. *P<0.05. miR,
microRNA; ASO, antisense; O.D., optical density; RT-qPCR,
reverse-transcription quantitative polymerase chain reaction; MTT,
methyl thiazolyl tetrazolium, FACS, fluorescence-activated cell
sorting. |
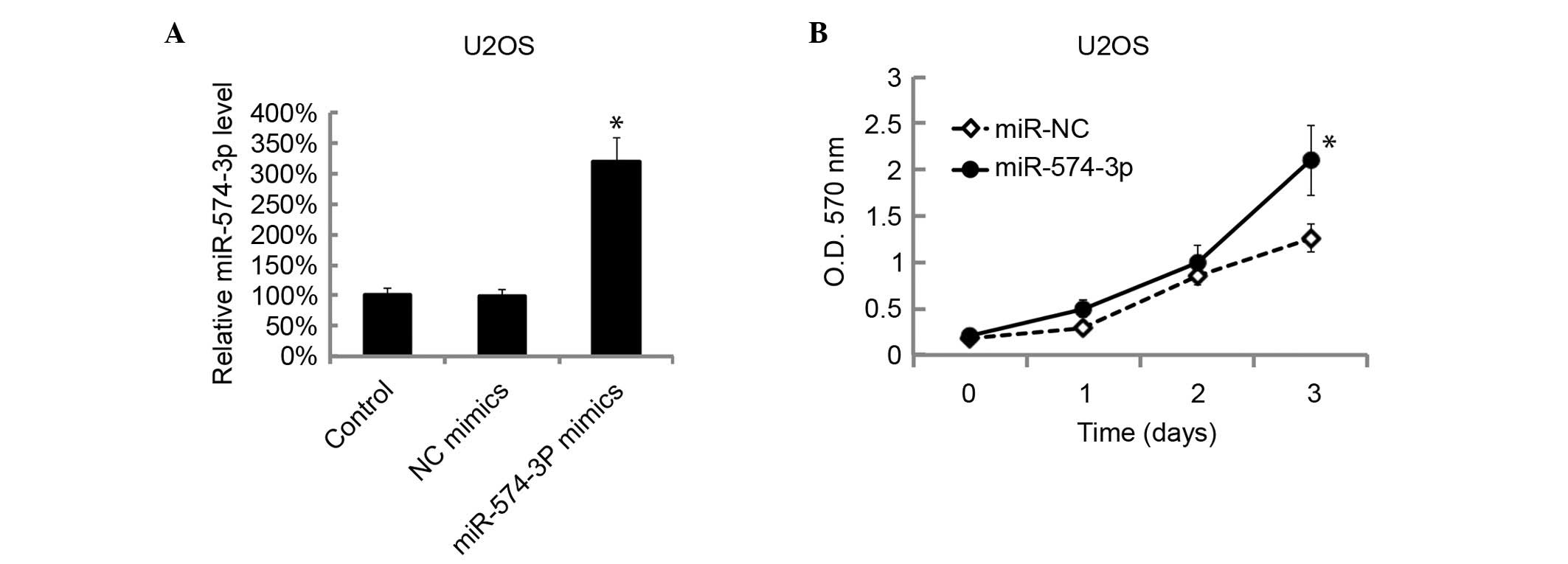
Overexpression of miR-574-3p promoted
cells growth
The level of miR-574-3p levels in U2OS cells was
overexpressed by transfection of miR-574-3p mimics. The miR-574-3p
levels in U2OS cells were assessed by RT-qPCR 48 h after
transfection. The miR-574-3p level in U2OS cells was significantly
overexpressed by miR-574-3p transfection with mimics (P=0.0093;
Fig. 3A). The cellular proliferation
was then examined by MTT analysis following miR-574-3p mimic
transfection. The overexpression of miR-574-3p significantly
promoted the cellular proliferation of U2OS (P=0.0115; Fig. 3B).
miR-574-3p directly targets the SMAD4
signaling pathway
To identify the potential targeted genes, SMAD4 was
predicted as a target gene of miR-574-3p by using TargetScan. SMAD4
is a unique common effector of tumor growth factor-β (TGF-β) and
bone morphogenetic protein signal pathways (16). SMAD4 is frequently mutated in human
carcinomas, leading to loss of growth inhibition by TGF-β (17). Thus, SMAD4 was chosen for additional
investigation. The 3′untranslated region (3′-UTR) and mutant 3′-UTR
of SMAD4 were used for the identification of target gene of
miR-574-3p (Fig. 4A). It was found
that miR-574-3p could markedly decrease the luciferase activity in
SMAD4 3′-UTR transfected cells while not in the mutant-transfected
cells (Fig. 4B). U2OS cells were then
transfected with miR-574-3p mimics. SMAD4 protein level was then
assessed by western blotting after 48 h. It was found that SMAD4
expression was also significantly inhibited in
miR-574-3p-expressing cells (P=0.0079; Fig. 4C). Cyclin-dependent kinase inhibitor 1
(CDKN1A), cyclin dependent kinase 4 inhibitor B (CDKN2B) and
inhibitor of DNA binding (ID3) are the SMAD4-dependent genes
(18). The present data showed that
CDKN1A, CDKN2B and ID3 were also significantly repressed in U2OS
cells by transfection with miR-574-3p mimics compared with the
negative control miRNA-transfected cells (P=0.0068; Fig. 4D).
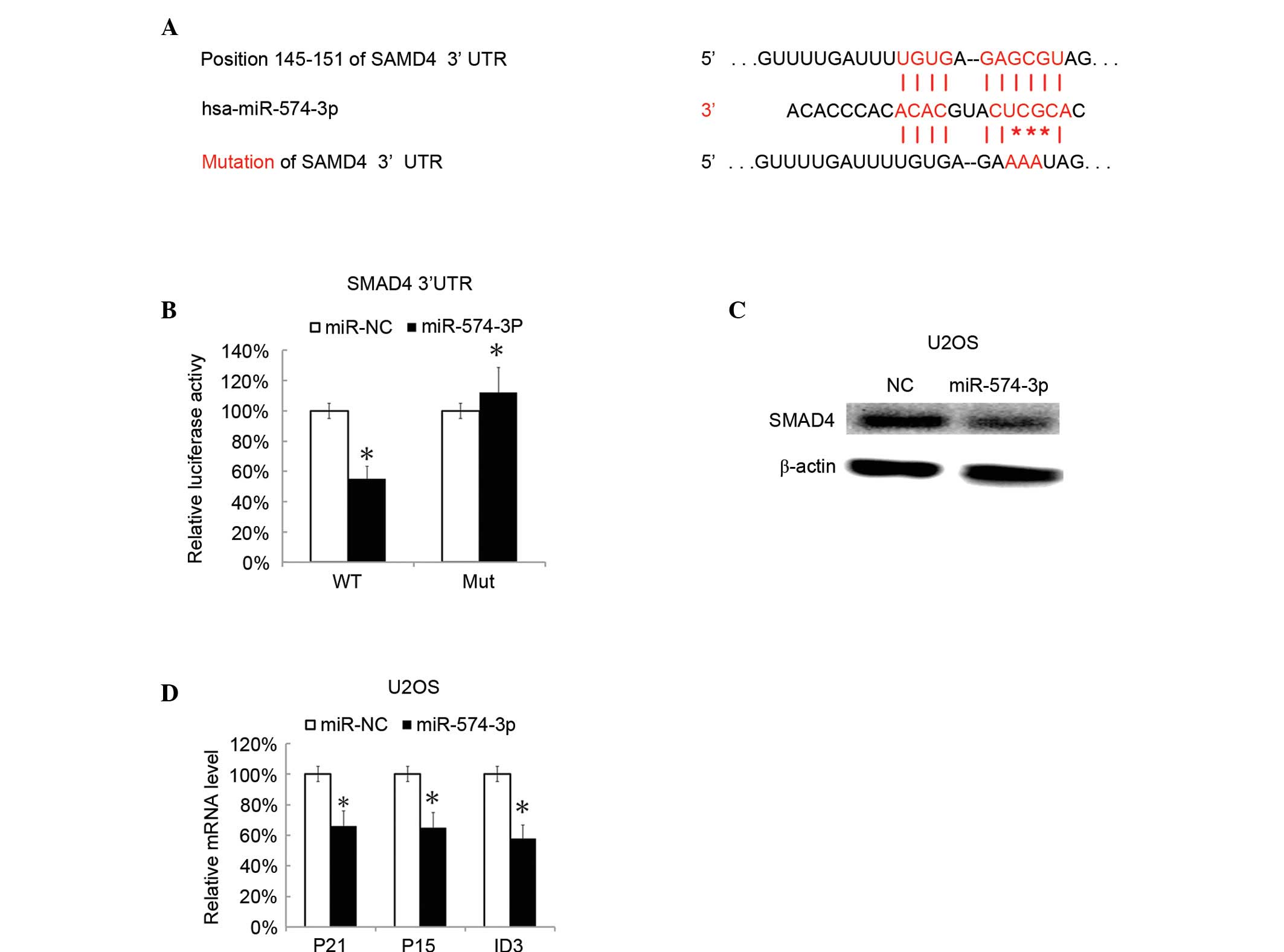 | Figure 4.SMAD4 was targeted by miR-574-3p.
Putative targeted genes were predicted by TargetScanHuman. (A) The
binding site of putative targeted gene, and mutated site of
miR-574-3p are shown. The RL reporter plasmids (RL-control,
RL-SMAD4, RL-mutated SMAD4) and miR-574-3p or miR-NC were
co-transfected into HEK293 cells, along with a firefly luciferase
reporter (pGL control) for normalization. (B) Luciferase activities
were determined following 48 h. The ratio of pGL activity vs. RL
luciferase activity in the miR-574-3p treated group was then
calculated and compared with the ratio in the miR-NC group (which
was arbitrarily defined as 100%). (C) Subsequent to 60 h after
transfection with miR-574-3p mimics SMAD4 protein levels were
assessed by western blotting. (D) Then, 60 h after transfection
with miR-574-3p mimics, the CDKN1A, CDKN2B and ID3 mRNA levels in
U2OS cells were assessed by RT-qPCR. All data are expressed as the
mean ± standard deviation, with experiments repeated three times.
*P<0.05. miR, microRNA; SMAD4, mothers against decapentaplegic
homolog 4; RL, Renilla luciferase; NC, negative control;
CDKN1A, cyclin-dependent kinase inhibitor 1; CDKN2B, cyclin
dependent kinase 4 inhibitor B; ID3, inhibitor of DNA binding;
RT-qPCR, reverse-transcription quantitative polymerase chain
reaction; WT, wild type; Mut, mutant. |
Overexpression of SMAD4 attenuated the
tumor promoting effects of miR-574-3p
The SMAD4 levels were investigated in osteosarcoma
tissues by RT-qPCR. It was found that osteosarcoma tissues showed
significantly decreased SMAD4 mRNA expression compared with
adjacent normal tissues (P=0.0058; Fig.
5A). Subsequently, the miR-574-3p levels and the SMAD4 levels
in 10 osteosarcoma tissue samples were analyzed, and it was found
that the miR-574-3p levels and the SMAD4 levels were negatively
correlated (Fig. 5B). By RT-qPCR, the
expression of SMAD4 target genes, CDKN1A, CDKN2B and ID2, were also
significantly decreased in osteosarcoma tissues (P=0.0093; Fig. 5C). U2OS cell were co-transfected with
SMAD4 overexpression plasmid, pc3.1-SMAD4 and miR-574-3p mimics.
The cell growth was evaluated by MTT analysis 48 h later. It was
found that that the tumor-promoting effect of miR-574-3p was
significantly inhibited by overexpression of SMAD4 (P=0.0124;
Fig. 5D).
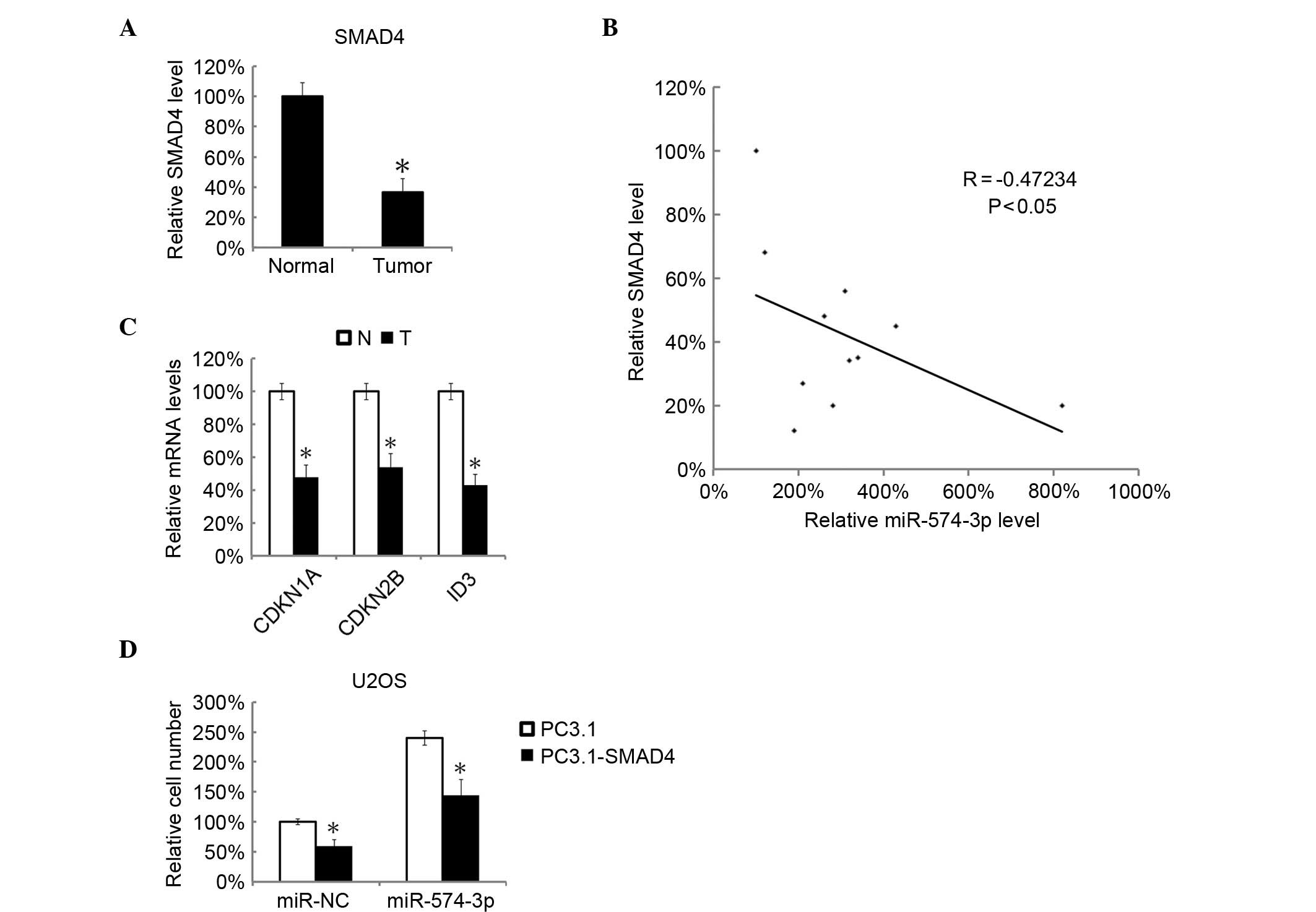 | Figure 5.miR-574-3p was negatively correlated
with the SMAD4 signaling pathway and overexpression of SMAD4
attenuated the tumor promoting effects of miR-574-3p. In total, 10
pairs of osteosarcoma and matched tumor-adjacent normal tissues
were analyzed by RT-qPCR for SMAD4 mRNA. (A) The mean level of
SMAD4 mRNA in tumor-adjacent normal tissues was arbitrarily defined
as 100%. (B) The correlation between SMAD4 mRNA and
miR-574-3p levels in the 10 osteosarcoma tissues was analyzed by
two-tailed Spearman's rank correlation coefficient analysis. (C)
The CDKN1A, CDKN2B and ID3 mRNA levels in 10 pairs of osteosarcoma
and matched tumor-adjacent normal tissues were analyzed by RT-qPCR.
The CDKN1A, CDKN2B and ID3 mRNA levels in tumor-adjacent normal
tissues were arbitrarily defined as 100%. U2OS cells were
co-transfected with pcDNA3.1-SMAD4 and miR-574-3p mimics; pcDNA3.1
empty plasmid and miR-NC were used as controls. (D) The relative
cell numbers were determined by MTT analysis 48 h subsequent to
transfection. The relative cell number in miR-NC and pcDNA3.1 empty
plasmid was arbitrarily defined as 100%. All data are mean ±
standard deviation with experiments repeated three times.
*P<0.05. miR, microRNA; NC, negative control; SMAD4, mothers
against decapentaplegic homolog 4; RT-qPCR, reverse-transcription
quantitative polymerase chain reaction; CDKN1A, cyclin-dependent
kinase inhibitor 1; CDKN2B, cyclin dependent kinase 4 inhibitor B;
ID3, inhibitor of DNA binding; N, normal; T, tumor. |
Discussion
miR-574 has been found to be upregulated in several
types of cancers, including lung cancer, bladder cancer and
prostate cancer (11–13,19). Foss
et al (17) also suggested
miR-574 as a serum biomarker of non-small cell lung cancer.
miR-574-3p negatively regulates Quaking 6/7 (Qki6/7), thus
affecting β-catenin/wingless (Wnt) signaling and the development of
colorectal cancer (12). Furthermore,
miR-574-3p was associated with various human cancers (19,20). In
the present study, it was demonstrated that miR-574-3p was
downregulated in human osteosarcoma tissues as well as osteosarcoma
U2OS, SAOS and MG63 cell lines. Inhibition of miR-574-3p by ASO
attenuated cell proliferation and resulted in the apoptosis in
osteosarcoma cells, while miR-574-3p mimics promoted the growth of
U2OS cells. Subsequently, SMAD4 was identified as a target of
miR-574-3p. SMAD4 overexpression could rescue the promoting effects
of miR-574-3p on cancer cell growth, which additionally supports
the theory that miR-574-3p may be associated with SMAD4.
miRNAs are critical regulators and biomarkers for
numerous types of cancers (8,21–24). There
are several miRNAs that have been found to be abnormal in human
osteosarcoma, including miR-141, miR-429, miR-451, miR-195, miR-183
miR-199a-3p, miR-127-3p and miR-376c (3,5,10,25–27). In
the present study, it was reported that miR-574-3p was highly
expressed in human osteosarcoma cells lines and tissues, and it was
indicated that miR-574-3p may be used as a biomarker for the
diagnosis of osteosarcoma.
The present study observed that miR-574-3p is
closely and positively associated with the proliferation of
osteosarcoma cells. Downregulation of miR-574-3p suppressed the
growth of U2OS cells and MG63 cells with the induction of
apoptosis, while overexpression of miR-574-3p enhanced the growth
of these cells. These data indicated that miR-574-3p plays an
oncogene-like function in osteosarcoma development. It has been
also reported that miR-574-3p targets Qki6/7 and affects colorectal
cancer through acting on the Wnt signaling pathway (12). In the present study, it was suggested
that SMAD4 is a direct target of miR-574-3p and the associated
genes CDKN1A, CDKN2B as well as ID3 were strongly suppressed by
miR-574-3p transfection. SMAD4 is often mutated in numerous cancers
and it acts as a tumor suppressor that is involved in the
regulation of the TGF-β signal transduction pathway, which
negatively regulates growth of epithelial cells and the
extracellular matrix (28–31). Therefore, the inhibition of SMAD4 and
its downstream proteins may be an important part of the mechanism
of miR-574-3p in osteosarcoma.
To investigate the importance of miR-574-3p-SMAD4
interaction in osteosarcoma, the current study determined the level
of SMAD4 and its associated genes (CDKN1A, CDKN2B and ID2) and
found that they were all decreased in osteosarcoma tissues. The
expression of miR-574-3p was inversely correlated with SMAD4
expression. Notably, SMAD4 overexpression rescued the
tumor-promoting effects of miR-574-3p, indicating that miR-574-3p
exerted an oncogenic effect, mainly through regulating the SMAD4
signaling pathway. However, a single miRNA could modulate >100
target genes (6), and the underlying
mechanism continues to require clarification in future studies.
In conclusion, the present study found that that
miR-574-3p is upregulated in human osteosarcoma, and plays an
oncogene-like role in human osteosarcoma through targeting the
tumor-suppressing gene SMAD4 and acting on the SMAD4 signaling
pathway. The current findings provide a novel target for the
diagnosis and treatment of human osteosarcoma.
References
|
1
|
de Oliveira JM Ferreira, Remédios C,
Oliveira H, Pinto P, Pinho F, Pinho S, Costa M and Santos C:
Sulforaphane induces Dna damage and mitotic abnormalities in human
osteosarcoma mg-63 cells: Correlation with cell cycle arrest and
apoptosis. Nutr Cancer. 66:325–334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Walter I, Wolfesberger B, Miller I, Mair
G, Burger S, Gallè B and Steinborn R: Human osteosarcoma cells
respond to sorafenib chemotherapy by downregulation of the tumor
progression factors S100A4, CXCR4 and the oncogene FOS. Oncol Rep.
31:1147–1156. 2014.PubMed/NCBI
|
|
3
|
Xu H, Mei Q, Shi L, Lu J, Zhao J and Fu Q:
Tumor-Suppressing effects of miR451 in human osteosarcoma. Cell
Biochem Biophys. 69:163–168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li R, Zhang W, Cui J, Shui W, Yin L, Wang
Y, Zhang H, Wang N, Wu N, Nan G, et al: Targeting BMP9-promoted
human osteosarcoma growth by inactivation of notch signaling. Curr
Cancer Drug Targets. 14:274–285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Liu Y, Wu S, Shi X, Li L, Zhao J
and Xu H: Tumor-suppressing effects of mir-429 on human
osteosarcoma. Cell Biochem Biophys. 70:215–224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chu Y, Zhu H, Lv L, Zhou Y and Huo J:
MiRNA s in oesophageal squamous cancer. Neth J Med. 71:69–75.
2013.PubMed/NCBI
|
|
7
|
Lai X, Wolkenhauer O and Vera J: Modeling
miRNA regulation in cancer signaling systems: miR-34a regulation of
the p53/Sirt1 signaling module. Methods Mol Biol. 880:87–108. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mo MH, Chen L, Fu Y, Wang W and Fu SW:
Cell-free circulating miRNA biomarkers in cancer. J Cancer.
3:432–448. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ahmed FE: miRNA as markers for the
diagnostic screening of colon cancer. Expert Rev Anticancer Ther.
14:463–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu H, Mei Q, Xiong C and Zhao J:
Tumor-suppressing effects of miR-141 in human osteosarcoma. Cell
Biochem Biophys. 69:319–325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu H, Liu X and Zhao J: Down-regulation of
miR-3928 promoted osteosarcoma growth. Cell Physiol Biochem.
33:1547–1556. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu X, Liu Y, Wu S, Shi X, Li L, Zhao J
and Xu H: Tumor-suppressing effects of miR-429 on human
osteosarcoma. Cell Biochem Biophys. 70:215–224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mei Q, Li F, Quan H, Liu Y and Xu H:
Busulfan inhibits growth of human osteosarcoma through miR-200
family microRNAs in vitro and in vivo. Cancer Sci. 105:755–762.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chiyomaru T, Yamamura S, Fukuhara S,
Hidaka H, Majid S, Saini S, Arora S, Deng G, Shahryari V, Chang I,
et al: Genistein up-regulates tumor suppressor microRNA-574-3p in
prostate cancer. PLoS One. 8:e589292013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji S, Ye G, Zhang J, Wang L, Wang T, Wang
Z, Zhang T, Wang G, Guo Z, Luo Y, et al: miR-574-5p negatively
regulates Qki6/7 to impact β-catenin/Wnt signalling and the
development of colorectal cancer. Gut. 62:716–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Foss KM, Sima C, Ugolini D, Neri M, Allen
KE and Weiss GJ: miR-1254 and miR-574-5p: Serum-based microRNA
biomarkers for early-stage non-small cell lung cancer. J Thorac
Oncol. 6:482–488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui J, Cheng Y, Zhang P, Sun M, Gao F, Liu
C and Cai J: down regulation of miR200c promotes radiation-induced
thymic lymphoma by targeting BMI1. J Cell Biochem. 115:1033–1042.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Y and Massagué J: Mechanisms of
TGF-beta signaling from cell membrane to the nucleus. Cell.
113:685–700. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schutte M: DPC4/SMAD4 gene alterations in
human cancer, and their functional implications. Ann Oncol.
10:(Suppl 4). 56–59. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kowanetz M, Valcourt U, Bergström R,
Heldin CH and Moustakas A: Id2 and Id3 define the potency of cell
proliferation and differentiation responses to transforming growth
factor beta and bone morphogenetic protein. Mol Cell Biol.
24:4241–4254. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su Y, Ni Z, Wang G, Cui J, Wei C, Wang J,
Yang Q, Xu Y and Li F: Aberrant expression of microRNAs in gastric
cancer and biological significance of miR-574-3p. Int
Immunopharmacol. 13:468–475. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meyers-Needham M, Ponnusamy S, Gencer S,
Jiang W, Thomas RJ, Senkal CE and Ogretmen B: Concerted functions
of HDAC1 and microRNA-574-5p repress alternatively spliced ceramide
synthase 1 expression in human cancer cells. EMBO Mol Med. 4:78–92.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang
B, Min L and Liu W: miRNA-200c increases the sensitivity of breast
cancer cells to doxorubicin through the suppression of
E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 7:1579–1584.
2013.PubMed/NCBI
|
|
25
|
Sun D, Layer R, Mueller AC, Cichewicz MA,
Negishi M, Paschal BM and Dutta A: Regulation of several
androgen-induced genes through the repression of the
miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells.
Oncogene. 33:1448–1457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martin EC, Bratton MR, Zhu Y, Rhodes LV,
Tilghman SL, Collins-Burow BM and Burow ME: Insulin-like growth
factor-1 signaling regulates miRNA expression in MCF-7 breast
cancer cell line. PLoS One. 7:e490672012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pignot G, Cizeron-Clairac G, Vacher S,
Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B,
Amsellem-Ouazana D and Bieche I: microRNA expression profile in a
large series of bladder tumors: Identification of a 3-miRNA
signature associated with aggressiveness of muscle-invasive bladder
cancer. Int J Cancer. 132:2479–2491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ouyang L, Liu P, Yang S, Ye S, Xu W and
Liu X: A three-plasma miRNA signature serves as novel biomarkers
for osteosarcoma. Med Oncol. 30:3402013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Novello C, Pazzaglia L, Cingolani C, Conti
A, Quattrini I, Manara MC, Tognon M, Picci P and Benassi MS: miRNA
expression profile in human osteosarcoma: Role of miR-1 and
miR-133b in proliferation and cell cycle control. Int J Oncol.
42:667–675. 2013.PubMed/NCBI
|
|
30
|
Maire G, Martin JW, Yoshimoto M,
Chilton-MacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu
M, Feng XH, Sawaya R, Medema RH, Hung MC and Huang S: Sustained
activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer
metastasis. J Clin Invest. 124:564–579. 2014. View Article : Google Scholar : PubMed/NCBI
|