Introduction
Recent studies have indicated that normal cellular
enzymatic activity may contribute to the genomic changes associated
with various neoplasias (1–3). Apolipoprotein B mRNA-editing enzymes
(APOBEC) comprise a family of enzymes that protect immune function
and are involved in mRNA editing; their cytosine deaminase activity
may also induce base substitutions in the genomes of malignancies
(1,2).
APOBEC3B is a cytosine deaminase that is responsible
for the deamination of cytosines in the genome of host cells,
producing cytosine to thymidine mutations (2). Recently studies have found that APOBEC3B
is overexpressed in several types of malignancy, resulting in
clusters of C>T mutations that are considered to be the
signature of APOBEC mutation in tumors. Analysis of whole-genome
and whole-exome sequencing data from various types of malignancy
have implicated APOBEC members in mRNA editing and the production
of cytosine mutation clusters in the development or progression of
numerous neoplastic processes, encompassing lung, breast, hepatic
and hematopoietic malignancies (4–9). APOBEC3B
overexpression has been identified in breast, head and neck cancers
(7–10). Although APOBEC3B expression has not
been studied in meningiomas, the presence of C>T point mutations
in some meningioma cases raises the possibility that APOBEC3B may
participate in the pathogenesis of at least a subset of meningiomas
(11).
The present study evaluated APOBEC3B expression in
addition to mutations in a series of normal leptomeninges, and
World Health Organization (WHO) grade I, II and III
meningiomas.
Materials and methods
Human leptomeningeal and meningioma
tissue
Frozen tissues from 5 fetal and 1 adult
leptomeninges and 38 meningiomas, including 17 WHO grade I, 17
grade II and 6 grade III meningiomas (12), were collected at the University of
Rochester Medical Center (Rochester, NY, USA), or obtained
primarily from the Cooperative Human Tissue Network (Philadelphia,
PA, USA), between January 2007 and December 2014, without
particular selection criteria (Table
I).
 | Table I.Summary of meningioma and
leptomeningeal tissues used in the present study. |
Table I.
Summary of meningioma and
leptomeningeal tissues used in the present study.
| A, Tissues used in
western blot analysis |
|---|
|
|---|
| Tissue subtype | n (%) | Mean patient age | Patient genders |
|---|
| Leptomeninges | 6 |
|
|
|
Fetal | 5 | – | U |
|
Adult | 1 | 40 years | M |
| WHO grade I
meningioma | 17 | 57 years | 14 F, 3 M |
|
Meningothelial | 7 (41) |
|
|
|
Transitional | 7 (41) |
|
|
|
Fibrous | 3 (18) |
|
|
| WHO grade II
meningioma | 15 | 53 years | 12 F, 3 M |
|
Meningothelial | 6 (40) |
|
|
|
Transitional | 3 (20) |
|
|
|
Fibrous | 5 (33) |
|
|
|
Microcystic | 1 (7) |
|
|
| WHO grade III
meningioma | 6 | 61 years | 4 F, 2 M |
|
Anaplastic | 3 (50) |
|
|
|
Transitional | 1 (17) |
|
|
|
Fibrous | 1 (17) |
|
|
|
Papillary | 1 (17) |
|
|
|
| B, Tissues used in
polymerase chain reaction analysis |
|
| Tissue subtype | n (%) | Mean patient age | Patient genders |
|
| Leptomeninges | 3 |
|
|
|
Fetal | 2 | 18 weeks | U |
|
Adult | 1 | 67 years | 1 F |
| WHO grade I
meningioma | 9 | 61 years | 7 F, 2 M |
|
Meningothelial | 6 (67) |
|
|
|
Transitional | 3 (33) |
|
|
| WHO grade II
meningioma | 10 | 60 years | 6 F, 4
M |
|
Meningothelial | 3 (30) |
|
|
|
Transitional | 1 (10) |
|
|
|
Fibrous | 4 (40) |
|
|
|
Secretory | 1 (10) |
|
|
|
Microcystic | 1 (10) |
|
|
| WHO grade III
meningioma | 4 | 64 years | 3 F, 1 M |
|
Anaplastic | 3 (75) |
|
|
|
Papillary | 1 (25) |
|
|
|
| C, Sources of
leptomeningeal and meningioma cells used for primary cultures |
|
| Type | Patient
age/gender | Location |
Classification/grade |
|
| LC1 | 20 weeks/U | Convexity | Leptomeninges |
| LC2 | 20 weeks/U | Convexity | Leptomeninges |
| LC3 | 22 weeks/U | Convexity | Leptomeninges |
| MC1 | 84 years/M | Right frontal | Meningothelial/I |
| MC2 | 65 years/F | Clivus | Meningothelial/I |
| MC3 | 80 years/M | Left frontal | Transitional/II |
Human leptomeningeal and meningioma
cell cultures
Primary leptomeningeal cultures were established
from two 20-week and one 22-week de-identified human fetuses ~5 h
after mortality from non-neurological disease. These were minced
and grown in petri dishes. Meningioma cell cultures were
established from fragments from the center of two WHO grade I and
one grade II meningiomas. Tissues were minced prior to plating in
flasks. These tissues along with the cerebrospinal fluid were
collected with the approval of the Institutional Review Board of
the University of Rochester. The cells were maintained in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum
(FBS; Gibco, Thermo Fisher Scientific, Inc.), which is insufficient
for the survival and proliferation of normal glial and endothelial
cells. The cells used were screened according to procedures
described previously, including the detection of a leptomeningeal
marker, epithelial membrane antigen (EMA) (13). For experiments, only early passages
(passages 2 to 5) were used of the meningioma cell cultures that
exhibited EMA immunoreactivity by immunocytochemistry (11). For immunocytochemical
characterization, meningioma cell cultures were plated onto 2-well
microscope slides (Nalgene NUNC International, Rochester, NY, USA)
for 1 day in DMEM with 10% FBS, prior to fixation in formalin.
Immunocytochemistry was performed using a mouse monoclonal antibody
against EMA (prediluted; Clone 29; #N1504; Dako, Carpenteria, CA,
USA) and visualized using the streptavidin-biotin-horseradish
peroxidase method (Dako). Extensive cytoplasmic immunoreactivity
was considered positive.
Human cerebrospinal fluid from
patients without neurological disease
As meningiomas are bathed in cerebrospinal fluid,
which has been demonstrated to affect meningioma cell proliferation
(13), its effects on APOBEC3B were
evaluated in the current study. Remnant, discarded lumbar
cerebrospinal fluid was collected at the University of Rochester
Medical Center and sent to the Department of Pathology between
March 2009 and September 2012. Samples from patients who were found
to be free of neurological disease, and who had a cytopathological
analysis showing no erythrocytes or increased lymphocytes were
classified as ‘normal’. In patients who had cerebrospinal fluid
collected as part of a staging protocol for a peripheral lymphoma,
only cerebrospinal fluid from those with no lymphoma cells or
abnormal numbers of lymphocytes by cytology and no neurological
disease were used. In some cases, due to limited volumes, multiple
samples were combined to achieve quantities sufficient for the
experimental design. Cerebrospinal fluid samples were initially
frozen at −17°C prior to storage at −80°C.
Western blot analysis of APOBEC3B
protein expression in human leptomeningeal and meningioma
tissues
For western blot analysis, meningioma lysates were
obtained by mechanical homogenization in Upstate RIPA Lysis Buffer
(EMD Millipore, Billerica, MA, USA) with 1:100 Protease Inhibitor
Cocktail (Sigma-Aldrich; EMD Millipore) then frozen at −85°C. The
tissue slurries were thawed on ice and vortexed vigorously, prior
to the removal of solids by centrifugation at 4°C. Protein
concentrations were quantified using a Bradford assay (Bio-Rad
Protein Assay; Bio-Rad Laboratories, Inc., Hercules, CA, USA), and
30–50 µg of protein from each sample was loaded and run on a 7.5%
acrylamide gel, before transfer to 0.45-µm nitrocellulose membrane.
The membrane was blocked for 1 h in 5% milk in Tris-Cl buffer with
Tween 20, then reacted with a polyclonal affinity-purified primary
antibody against APOBEC3B (#sc-130955; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) overnight at 4°C. This was followed by
horseradish peroxidase-conjugated secondary antibody treatment
(#170-6515; Bio-Rad Laboratories, Inc.). Detection was achieved
using Clarity Western ECL Substrate (Bio-Rad Laboratories, Inc.)
and imaging was performed with Bio-Rad ChemiDoc instrumentation and
Image Lab software (Bio-Rad Laboratories, Inc.). Loading was
assessed using a polyclonal rabbit antibody against actin
(dilution, 1:3000; #4967; Cell Signaling Technology, Inc., Danvers,
MA, USA); to evaluate the results, the APOBEC3B band intensity was
normalized relative to that of actin.
Analysis of APOBEC3B mRNA in
leptomeningeal tissue and meningioma tumors
RNA was isolated from 2 fetal and 1 adult
leptomeninges, 9 WHO grade I, 10 WHO grade II and 4 WHO grade III
meningiomas using an RNeasy Plus universal (#7304; Qiagen, Inc.,
Valencia, CA, USA) according to manufacturers specifications.
Reverse transcription (RT) was performed in a 20-µl reaction
mixture containing of 1 µg of RNA, using an iScript cDNA Synthesis
Kit (Bio-Rad Laboratories, Inc.); the RT conditions were 5 min at
25°C, 30 min at 42°C, 5 min at 85°C, and an indefinite hold at 4°C.
The resulting cDNA was diluted to 200 µl with water.
Quantitative polymerase chain reaction (qPCR) for
APOBEC3B was performed in triplicate using primers for APOBEC3B
(forward, 5′-GACCCTTTGGTCCTTCGAC-3′; reverse,
5′-GCACAGCCCCAGGAGAAG-3′) and the RPL13A control gene (forward
5′-AGATGGCGGAGGTGCAG-3′ and reverse 5′-GCCAGAAATGTTGATGCCTT-3′).
The qPCR reaction mixture was composed of 1X iTaq Universal SYBR
Green Supermix (Bio-Rad Laboratories, Inc.), 1 µM primer mix, and
4.9 µl of diluted cDNA in a total volume of 10 µl. Reactions were
performed on the Bio-Rad CFX Connect Real-Time System using the
default protocol CFX_2StepAmp+Melt, as follows: 95°C for 3 min,
followed by 40 cycles of 95°C for 10 sec and 55°C for 30 sec, and
95°C for 10 sec. Data was collected using Bio-Rad CFX Manager 3.0
software (Bio-Rad Laboratories, Inc.). Relative quantification was
performed using the 2−ΔΔCq calculation method
(14).
Sequencing of APOBEC3B DNA in
leptomeningeal tissue and meningioma tumors
DNA was isolated from frozen meningioma tissue using
the Qiagen DNeasy Blood and Tissue Kit according to the
manufacturer's guidelines, and stored at −80°C. Sequencing was
performed using a TruSeq Amplicon-Cancer Panel (Illumina, Inc., San
Diego, CA, USA). Sequences and controls were compared against the
known sequence.
Effects of platelet-derived growth
factor-BB (PDGF-BB) and cerebrospinal fluid on APOBEC3B protein
expression
PDGF-BB and cerebrospinal fluid were previously
demonstrated to influence leptomeningeal and meningioma cell
proliferation (13). Consequently,
confluent cells from 3 primary fetal leptomeningeal cell cultures
(gestational ages 20, 20 and 22 weeks), 2 WHO grade I and 2 WHO
grade II primary meningioma cultures were serum-deprived overnight
prior to treatment with serum-free DMEM, DMEM containing 10 ng/ml
PDGF-BB, or pooled cerebrospinal fluid from patients, for 72 h.
Lysates of the cells were then analyzed by western blotting, as
follows. Meningioma lysates were homogenized in RIPA Lysis Buffer
as described above. Following quantification of protein
concentrations, 10 µg protein from each sample was loaded and run
on a 7.5% acrylamide gel, then transferred to a 0.45-µm
nitrocellulose membrane. The membrane was blocked for 1 h in 5%
milk in Tris-Cl buffer with Tween 20, then reacted with the
affinity-purified primary antibody overnight at 4°C. This was
followed by horseradish peroxidase-conjugated secondary antibody
treatment. Detection was achieved with Western Lightning reagent
(PerkinElmer, Inc., Waltham, MA, USA) on Kodak Xomat film. All
western blots were subsequently repeated and quantified with
Clarity Western ECL substrate (Bio-Rad Laboratories, Inc.) and
Chemidoc software.
Results
APOBEC3B protein expression in
leptomeninges and meningioma
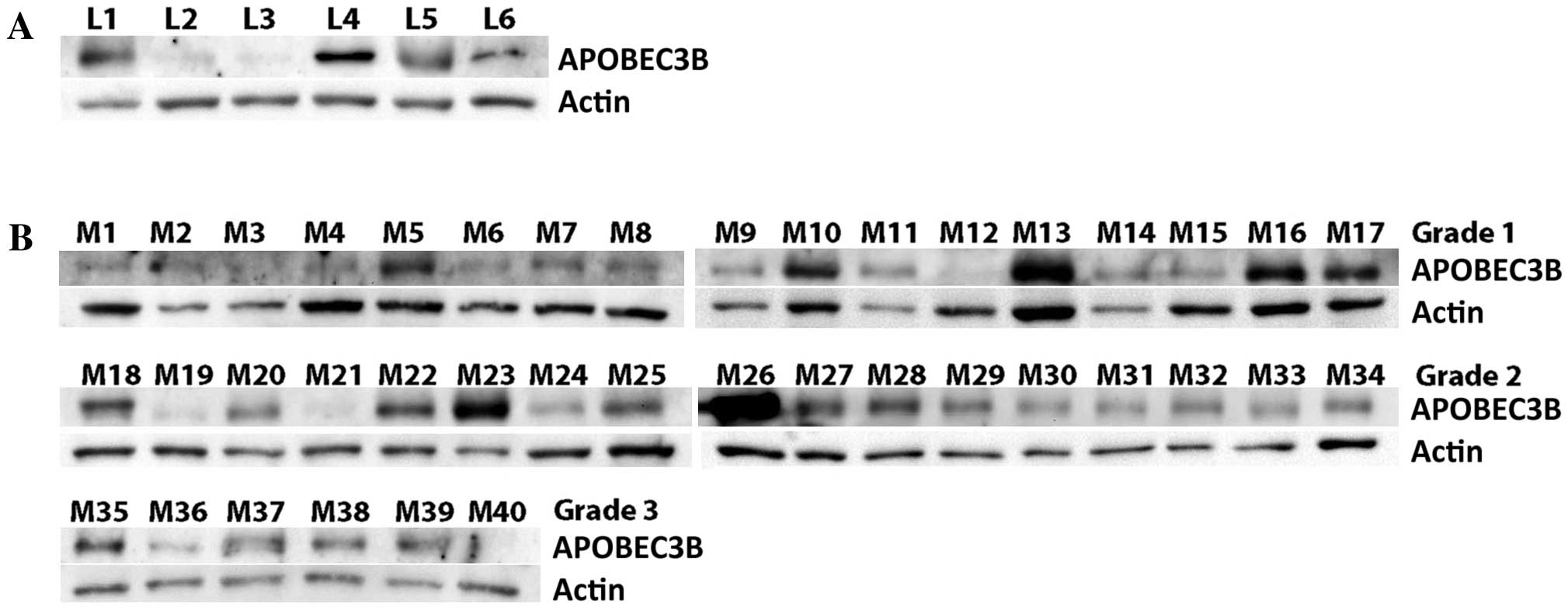
As shown in Fig. 1A,
APOBEC3B was detected in the 16-, 17-, 20-, 22- and 23-week fetal
and the adult leptomeningeal tissues. In the meningiomas, APOBEC3B
was detected in 15 of 17 WHO grade I meningiomas (Fig 1B). APOBEC3B was also detected in all 17
WHO grade II and 5 of 6 grade III tumors (Fig. 1B).
Analysis of APOBEC3B mRNA in
leptomeningeal tissue and meningioma tumors
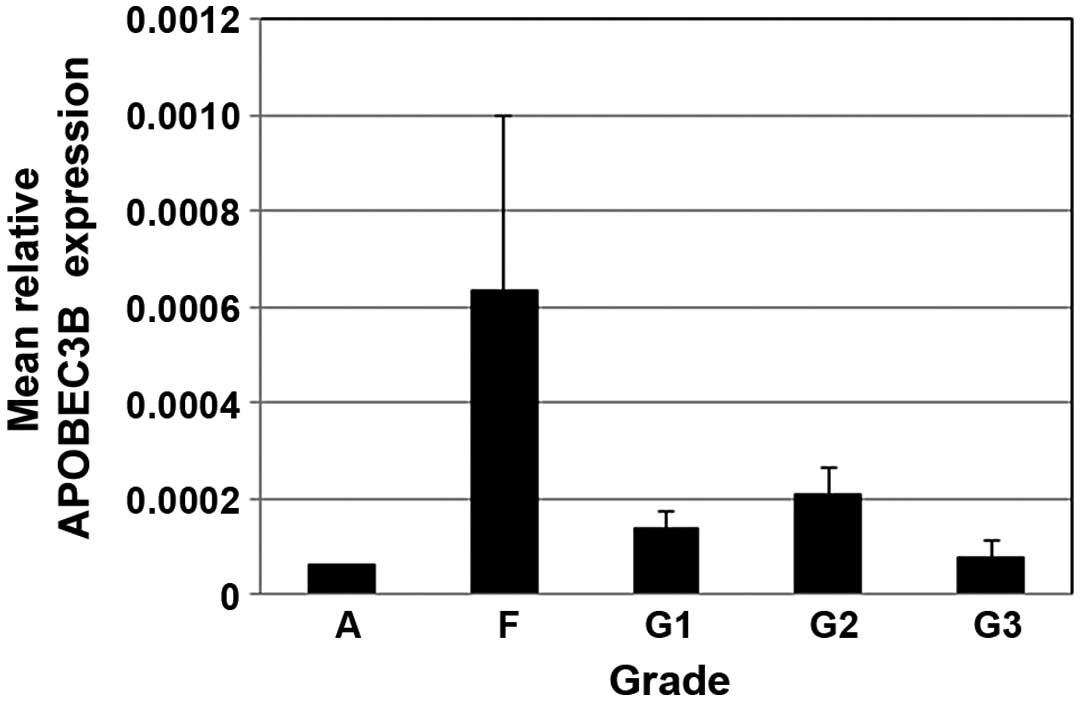
qPCR was used to assess the expression levels of
APOBEC3B, which were calculated relative to RPL13A for each
specimen and normalized to expression in the adult meninges
specimen. The relative APOBEC3B expression in fetal leptomeninges
was 4-fold higher than that in normal adult tissue. APOBEC3B
expression levels tended to be lower in grade III compared to grade
I and II tumors, but were not markedly different (Fig. 2).
Sequencing of APOBEC3B DNA in
leptomeningeal tissue and meningioma tumors
No point mutations were detected in APOBEC3B in the
2 WHO grade I, 3 grade II and 3 grade III meningiomas.
Effects of PDGF-BB and cerebrospinal
fluid on APOBEC3B protein expression
The 20- and 22-week fetal leptomeningeal cells
treated with PDGF-BB or human cerebrospinal fluid demonstrated no
changes in APOBEC3B protein expression. Additionally, in the WHO
grade I and II meningioma cell cultures, these treatments had no
effect (data not shown).
Discussion
APOBEC3B mRNA has been demonstrated to be
overexpressed in certain epithelial malignancies, including a high
proportion of breast, ovarian and lung carcinomas (5,15,16), and has been shown to promote the
development of malignancy (7–10). This is considered to be due to an
increased transition from dC to dT and the resultant increased
mutation frequency (5). These
mutations were commonly found in oncogenes transcribed in tumor
cells (17); for example, in lymphoma
cell lines, overexpression of APOBEC3B produced an increase in dC
to dT mutations in c-Myc (18). In
the present study, it was shown that APOBEC3B was present in all of
the fetal leptomeninges, 88% of WHO grade I, 100% of grade II and
83% of grade III meningiomas tested, without differences between
grades. PCR revealed no overexpression and sequencing revealed no
mutation in APOBEC3B. These findings suggest that, in contrast to
several other epithelial malignancies, the overexpression and
C>T point mutations in the genome of meningiomas may not be
critical to the pathogenesis of meningiomas. Nonetheless, the
number of WHO grade III meningiomas available for analysis was
limited, and we therefore cannot exclude the possibility that some
subtypes of grade III meningiomas may exhibit overexpression.
High APOBEC expression was identified in a few,
particularly WHO grade II, meningiomas. Like the majority of
meningiomas in this study, these were from tumors in female
patients. However, these were distributed amongst histological
subtypes with no distinct predilection for site. Nonetheless, the
number from any location was limited.
Based on embryological studies, other deaminase
DNA-editing enzyme families may be more important in the
pathogenesis of meningioma. For example, while the the APOBEC
family is important in embryogenesis, the adenosine deaminase
acting on RNA (ADAR) family is more important in brain development
(19). Nonetheless, the current study
identified no definite changes in APOBEC expression among the
meningiomas in this series.
References
|
1
|
Avesson L and Barry G: The emerging role
of RNA and DNA editing in cancer. Biochim Biophys Acta.
1845:308–316. 2014.PubMed/NCBI
|
|
2
|
Kuong KJ and Loeb LA: APOBEC3B mutagenesis
in cancer. Nat Genet. 45:964–965. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Benne R, Van den Burg J, Brakenhoff JP,
Sloof P, Van Boom JH and Tromp MC: Major transcript of the
frameshifted coxII gene from trypanosome mitochondria contains four
nucleotides that are not encoded in the DNA. Cell. 46:819–826.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burns MB, Lackey L, Carpenter MA, Rathore
A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N,
Nikas JB, et al: APOBEC3B is an enzymatic source of mutation in
breast cancer. Nature. 494:366–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burns MB, Temiz NA and Harris RS: Evidence
for APOBEC3B in multiple human cancers. Nat Genet. 45:977–983.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lada AG, Dhar A, Boissy RJ, Hirano M,
Rubel AA, Rogozin IB and Pavlov YI: AID/APOBEC cytosine deaminase
induces genome-wide kataegis. Biol Direct. 7:472012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor BJ, Nik-Zainal S, Wu YL, Stebbings
LA, Raine K, Campbell PJ, Rada C, Stratton MR and Neuberger MS: DNA
deaminases induce break-associated mutation showers with
implication of APOBEC3B and 3A in breast cancer kataegis. ELife.
2:e005342013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roberts SA, Lawrence MS, Klimczak LJ,
Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL,
Saksena G, et al: An APOBEC cytadine deaminase pattern is
widespread in human cancers. Nat Genet. 45:970–976. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Okuyama S, Marusawa H, Matsumoto T, Ueda
Y, Matsumoto Y, Endo Y, Takai A and Chiba T: Excessive activity of
Apolipoprotein B mRNA editing enzyme catalytic polypeptide 2
(APOBEC2) contributes to liver and lung tumorigenesis. Int J
Cancer. 130:1294–1301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jia P, Pao W and Zhao Z: Patterns and
processes of somatic mutations in nine major cancers. BMC Med
Genomics. 7:112014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goutagny S, Yang HW, Zuchman-Rossi J, Chan
J, Dreyfuss JM, Park PJ, Black PM, Giovanni M, Carroll RS and
Kalamarides M: Genomic profiling reveals alternative genetic
pathways of meningioma malignant progression dependent on the
underlying NF2 status. Clin Cancer Res. 16:4155–4164. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Perry A, Louis DN, Scheithauer BW, Budka H
and von Deimling A: MeningiomasTumours of the Nervous System. Louis
DN, Ohgaki H, Wiestler OD and Cavenee WK: WHO Press; Geneva,
Switzerland: pp. 164–172. 2007
|
|
13
|
Johnson MD, O'Connell M, Facik M, Maurer
P, Jahromi B and Pilcher W: Cerebrospinal fluid stimulates
leptomeningeal and meningioma cell proliferation and activation of
STAT3. J Neurooncol. 107:121–131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leonard B, Hart SN, Burns MB, Carpenter
MA, Temiz NA, Rathore A, Vogel RI, Nikas JB, Law EK, Brown WL, et
al: APOBEC3B upregulation and genomic mutation patterns in serous
ovarian carcinoma. Cancer Res. 73:7222–7231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sasaki H, Suzuki A, Tatematsu T, Shitara
M, Hikosaka Y, Okuda K, Moriyama S, Yano M and Fujii Y: APOBEC3B
gene overexpression in non-small-cell lung cancer. Biomed Rep.
2:392–395. 2014.PubMed/NCBI
|
|
17
|
Shinohara M, Io K, Shindo K, Matsui M,
Sakamoto T, Tada K, Kobayashi M, Kadowaki N and Takaori-Kondo A:
APOBEC3B can impair genomic stability by inducing base
substitutions in genomic DNA in human cells. Sci Rep. 2:8062012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rebhandal S, Huemer M, Gassner FJ,
Zaborsky N, Hebenstreit D, Catakovic K, Grössinger EM, Greil R and
Geisberger R: APOBEC3 signature mutations in in chronic lymphocytic
leukemia. Leukemia. 28:1929–1932. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Avesson L and Barry G: The emerging role
of RNA and DNA editing in cancer. Biochim and Biophys Acta.
1845:308–316. 2014.
|