Introduction
Colorectal cancer (CRC) is the third most common
type of cancer and the leading cause of cancer-associated mortality
in men and women in the United States (1). The same trends in incidence and
mortality rates are present in China, with its incidence rapidly
increasing by 4.2% each year (2).
Metastases is the predominant cause of cancer-associated mortality.
The most important CRC prognostic factor in clinics is the American
Joint Committee on Cancer tumor-node-metastasis (TNM) staging,
which is determined by the depth of invasion, the involvement of
pericolorectal lymph nodes and distant metastasis (3). It is generally considered that patients
with stage II and III CRC have favorable prognoses. However, data
have shown that the rate of recurrence and metastasis for these
patients is as high as 30% (4). In
previous years, several molecular factors have been examined as
prognostic and predictive factors for CRC, including small mothers
against decapentaplegic 4, BRAF and extracellular signal-regulated
kinase 1/2 (5,6). However, efficient clinical predictive
markers to guide the screening of patients with CRC at high risk of
recurrence and metastasis remain to be elucidated. Therefore, it is
essential to identify novel prognostic and predictive factors for
CRC, other than the TNM stage, in order to minimize adverse effects
and maximize therapeutic effects in treating patients with CRC.
The development of CRC is a complex process based on
the accumulation of several genetic factors, including alterations
in oncogenes, tumor suppressor genes, and genes associated with DNA
damage and repair (7). The Nm23-H1
gene is a metastasis suppressor gene, the low expression of which
can promote tumor occurrence and metastasis during tumor
progression (8). As Nm23-H1 was
originally identified as a metastasis suppressor protein, its
expression has been correlated with tumor metastatic potential in
various types of human carcinoma, primarily in ductal breast cancer
(9) and CRC (7). Low expression levels of Nm23-H1 have
been associated with poor prognosis in gastric adenocarcinoma,
breast cancer, hepatocellular carcinoma and ovarian carcinoma
(9–12). p53 is a transcription factor involved
in regulating cell cycle arrest and apoptosis in response to DNA
damage and cellular stress, and is thereby critical in protecting
cells from malignant transformation (13,14). It
has been shown that analysis of the expression of p53 offers
considerable promise for accurately predicting recurrence and
progression rates in patients with tumors, including gastric cancer
(15) and breast cancer (16). Although the value of the expression of
p53 in the prognosis of patients with CRC has been widely
investigated (17,18), data analysis on the use of the
expression of p53 for long-term survival rate prediction in
patients with CRC is limited (17).
The present study aimed to investigate the
expression levels of Nm23-H1 and p53 in stage II and III CRC
tissues, and examine their association with the clinicopathological
features of the patients and tumors. The effect of the combined
detection of Nm23-H1 and p53 on the survival rates of patients with
CRC was also evaluated.
Materials and methods
Patients and specimens
A total of 110 paraffin-embedded samples were
collected from patients with CRC who underwent surgery at the First
Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China)
between 2001 and 2006. Medical records of patients enrolled in the
study were retrospectively analyzed. Pathological diagnoses were
classified in accordance with the World Health Organization (WHO)
criteria (19), and staging was
performed according to the American Joint Committee on Cancer TNM
classification (20,21). In addition, 53 cases of
paraffin-embedded normal colorectal specimens were included from
patients who underwent colorectal biopsy by colonoscopy. The
present study was approved by the Ethics Committee of the First
affiliated Hospital of Xi'an Jiaotong University.
Among the 110 patients with CRC, 58 (52.73%) were
men and 52 (47.27%) were women; 44 (40.00%) patients were <60
years old and 66 (60.00%) were >60 years old. In terms of
staging, 56 (50.9%) patients were in stage II and 54 (49.1%)
patients were in stage III. Regarding site, 65 (59.09%) tumors were
located in the colon and 45 (40.91%) tumors were located in the
rectum, of which 37 (33.63%) tumors had a diameter <5 cm
(including 5 cm) and 73 (66.36%) tumors >5 cm. According to
histological grade, 93 (84.55%) tumors were well to moderately
differentiated, whereas 17 (15.45%) tumors were poorly
differentiated. According to the WHO classification of CRC, 92
(83.64%) tumors were classified as adenocarcinomas, 12 (10.91%)
tumors were classified as mucinous carcinomas, five (4.55%) tumors
were classified as undifferentiated carcinomas, and one case was
designated as unclear. According to the TNM classification, 48
(43.64%) tumors exhibited T1-3 infiltration, and 62 (56.36%) tumors
exhibited T4 infiltration. In addition, 56 (50.91%) cases were
without lymphatic metastasis, whereas 54 (49.09%) cases exhibited
lymphatic metastasis, including 41 cases with <12 lymph nodes in
biopsy (41/54) and 13 cases with >12 (13/54) lymph nodes in
biopsy. In the control group, there were 19 women and 34 men, aged
between 32 and 79 years with a median age of 59 years.
Immunohistochemical analysis
Immunohistochemical staining for p53 and Nm23-H1 was
performed using a standard avidin-biotin complex method. All
specimens were fixed in 10% neutral-buffered formalin overnight at
room temperature, embedded in paraffin and cut into 4-µm-thick
sections. Subsequently, the sections were deparaffinized in xylene
baths and rehydrated with a series of ethanol solutions for 5 min
each. A microwave was used for antigen retrieval for 30 min in 0.01
M of sodium citrate. Following blocking of endogenous peroxidase
activity with 5% hydrogen peroxidase for 10 min, incubation with
the primary antibody was performed for 2 h at room temperature.
Mouse monoclonal antibody against p53 (sc-47698; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) was used at 1:500
dilution, and a mouse monoclonal antibody against Nm23-H1
(sc-514515; Santa Cruz Biotechnology, Inc.) was used at 1:100
dilution. The sections were then incubated with horseradish
peroxidase-conjugated secondary antibody (SP-9002; Zhongshan Golden
Bridge Biotechnology, Peking, China) at room temperature for 30
min. The tissue sections were then washed in tris-buffered saline
for 10 min, with 3-amino-9-ethylcarbazole used as a chromogen, and
hematoxylin counterstaining was performed.
Under a light microscope (Olympus BX53; Olympus
Corporation, Tokyo, Japan), p53 protein staining was observed to be
localized in the nucleus, and Nm23-H1 protein was primarily
expressed in the cytoplasm. Positive staining was visible as
brown-yellow nuclear/cytoplasmic staining for the respective
antibody. Antigen expression was evaluated in a semi-quantitative
manner. Immunoreactivity was scored based on the immunoreactive
cell percentage and staining intensity of each slide in each low
power field of three randomly selected microscopic fields per slide
(magnification, ×100). The immunoreactive cell percentage was
defined as follows: Staining index 0, tissue with no staining;
staining index 1, tissue with faint or moderate staining in <25%
of tumor cells; staining index 2, tissue with moderate or marked
staining in 25–75% of tumor cells; staining index 3, tissue with
marked staining in >75% of tumor cells. In addition, the
staining intensity, compared with the background, was defined as
follows: 0, colorless; 1, cream-colored; 2, brown-yellow; 3, tan.
The mean product of these two indices from three fields was defined
as the final score: 0–2 (−), 3–4 (+), 5–7 (++), 8–9 (+++).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Tissues were cut and incubated with
diethylpyrocarbonate. After full grinding, the tissue suspension
obtained through the strainer was collected and used for RNA
extraction. Total RNA was extracted from the frozen tumor and
normal colorectal tissue suspensions using an RNA fast 200 kit
(Fastagen Biotechnology, Co., Ltd., Shanghai, China). The RT
reaction was performed using a PrimeScript RT Reagent kit (Takara
Bio, Inc., Otsu, Japan). Subsequently, SYBR Premix EX Taq™II
(Takara Bio, Inc,) was used to perform qPCR analysis, according to
the manufacturer's protocols, and the results were detected using a
Prism 7500 real-time PCR detection system (Applied Biosystems;
Thermo Fisher Scientific, Waltham, MA, USA).). The reaction
conditions were as follows: 94°C for 3 min, followed by 25 cycles
of 94°C for 30 sec, 59.6°C for 30 sec and 72°C for 30 sec.
Oligonucleotide primers for p53, Nm23-H1 and GAPDH were synthesized
by Sangon (Sangon Biotech Co., Ltd., Shanghai, China). The primer
sequences were as follows: p53 forward,
5′-CCTCAGCATCTTATCCGAGTGG-3′ and reverse,
5′-TGGATGGTGGTACAGTCAGAGC-3′; Nm23-H1 forward,
5′-TTGACCGGGGTAGAGAACTC-3′ and reverse,
5′-TCTCAGTACTTCCCGTGACC-3-′; and GAPDH forward,
5′-AGGTCCACCACTGACACGTT-3′ and reverse, 5′-GCCTCAAGATCAGCAAT-3′.
GAPDH was used as an internal reference gene. The relative mRNA
expression levels were calculated using the delta-delta
quantification cycle method, as follows: Delta Ct =
CtTarget - CtGAPDH. (22).
Follow-up
The patients with CRC enrolled in the present study
were followed up at the outpatient clinic, or by telephone call or
letter. Follow-up was performed once every 3 months in the first
year following surgery, and once every 6 months subsequently. As of
December 2011, the follow-up duration for the 110 cases ranged
between 4 and 102 months, with a median duration of 72 months.
Statistical analysis
Statistical analysis was performed using SPSS
software version 17.0 (SPSS, Inc. Chicago, USA). Data are presented
as the mean ± standard deviation. Enumeration data were analyzed
using the χ2-test. Student's t-test was used for
comparison between two groups. Bonferroni correction was used in
multiple comparisons. Survival analysis was performed using
Kaplan-Meier curves and associated log-rank tests. Correlation
analysis was assessed by Spearman's test for ranked data. P<0.05
was considered to indicate a statistically significant
difference
Results
Protein expression of Nm23-H1 and p53
in CRC and normal colon tissues
Positive Nm23-H1 immunostaining was detected in
57.3% (63/110) of the CRC cases (Fig.
1A-C), and in 90.6% (48/53) of the normal tissue samples
(Fig. 1D). The expression levels of
Nm23-H1 in the CRC tissues were significantly lower, compared with
those in the normal tissues (χ2=18.249; P<0.001). In
addition, a significant correlation was observed between lymph node
metastasis and the expression of Nm23-H1 (χ2=11.847,
P=0.001). CRCs with lymph node metastasis stained positively for
Nm23-H1 in 40.7% (22/54) of the cases, whereas the CRC samples
without lymph node metastasis stained positively for Nm23-H1 in
73.2% (41/56) of the cases. A significant difference was also
observed in Nm23-H1 staining according to the WHO classification of
the tumors. It was found that 63.0% (58/92) of the adenocarcinoma
tissues, 33.3% (4/12) of the mucinous carcinoma tissues and 20.0%
(1/5) of the undifferentiated carcinoma tissues stained positively
for Nm23-H1 (χ2=6.911; P=0.032). However, no
correlations were found between the protein expression of Nm23-H1
and the other clinicopathologic features of the patients and
tumors, including gender (P=0.492), age (P=0.208), tumor location
(P=0.487), tumor size (P=0.741), grade (P=0.500) and invasive depth
(P=0.081). The results of these analyses are presented in Table I.
 | Table I.Association between expression levels
of Nm23-H1 and p53 and clinical pathological features of patients
with colorectal cancer. |
Table I.
Association between expression levels
of Nm23-H1 and p53 and clinical pathological features of patients
with colorectal cancer.
| Variable | n | Nm23-H1-positive rate
(%) | χ2
(P-value) | p53-positive rate
(%) | χ2
(P-value) |
|---|
| Gender |
|
| 0.473 (0.492) |
| 0.006 (0.938) |
| Male | 58 | 60.3 |
| 72.4 |
|
|
Female | 52 | 53.8 |
| 73.1 |
|
| Age (years) |
|
| 1.585 (0.208) |
| 1.719 (0.190) |
|
≤60 | 44 | 50.0 |
| 79.5 |
|
|
>60 | 66 | 62.1 |
| 68.2 |
|
| Tumor location |
|
| 0.483 (0.487) |
| 0.014 (0.905) |
|
Colon | 65 | 60.0 |
| 72.3 |
|
|
Rectum | 45 | 53.3 |
| 73.3 |
|
| Tumor size
(cm) |
|
| 0.109 (0.741) |
| 0.244 (0.621) |
| ≤5 | 37 | 54.1 |
| 71.2 |
|
|
>5 | 73 | 58.9 |
| 75.7 |
|
| Gradea |
|
| 0.454 (0.500) |
| 0.652 (0.102) |
|
I–II | 93 | 55.9 |
| 74.2 |
|
|
III | 17 | 64.7 |
| 64.7 |
|
| Pathological
patternb |
|
| 6.911
(0.032)c |
| 6.203 (0.102) |
|
Adenocarcinoma | 92 | 63.0 |
| 78.1 |
|
|
Mucinous carcinoma | 12 | 33.3 |
| 54.2 |
|
|
Undifferentiated
carcinoma | 5 | 20.0 |
| 81.8 |
|
| Invasive depth |
|
| 3.046 (0.081) |
| 0.002 (0.969) |
|
T1-3 | 48 | 47.9 |
| 72.9 |
|
| T4 | 62 | 64.5 |
| 72.6 |
|
| Lymphatic
metastasis |
|
| 11.847
(0.001)c |
| 0.547 (0.459) |
| No | 56 | 73.2 |
| 69.6 |
|
|
Yes | 54 | 40.7 |
| 75.9 |
|
The protein expression levels of p53 in CRC tissues
varied. Positive staining was observed as brown granules,
predominantly localized in the nucleus. Positive p53 staining was
observed in 80 CRC cases (72.7%; Fig.
1E-G), whereas 30 of the normal tumors were found to be
negative for the protein expression of p53 (Fig. 1H). Compared with this observation for
CRC tumors, only three cases were positive for the expression of
p53 in the 53 normal controls (5.7%). These results showed that the
expression levels of p53 in CRC tissues were significantly higher,
compared with those in normal tissues (χ2=23.940;
P<0.001). No significant correlation was found between the
expression of p53 and the analyzed clinicopathological features of
the patients and their tumors, including gender (P=0.938), age
(P=0.190), tumor location (P=0.905), tumor size (P=0.621), grade
(P=0.102), pathological pattern (P=0.102), invasive depth (P=0.969)
and lymphatic metastasis (P=0.459). The results of these analyses
are presented in Table I.
Correlation between the protein
expression of Nm23-H1 and p53 in CRC tissues
The Nm23-H1-positive CRC cases were divided based on
staining intensity: 19 cases were 1+, 27 cases were 2+ and 17 cases
were 3+. The 80 p53-positive cases were also divided based on
staining intensity: 27 cases were 1+, 30 cases were 2+, and 23
cases were 3+ (Table II). The
correlation between the protein expression of Nm23-H1 and p53 was
analyzed using Spearman's test, and the results revealed that the
protein expression of p53 was negatively correlated with the
protein expression of Nm23-H1 (rs=−0.400;
P<0.001).
 | Table II.Correlation of protein expression of
Nm23-H1 and p53 in 110 colorecatal cancer tissue samples. |
Table II.
Correlation of protein expression of
Nm23-H1 and p53 in 110 colorecatal cancer tissue samples.
|
| Nm23-H1
expression |
|
|---|
|
|
|
|
|---|
| p53 expression | Negative | 1+ | 2+ | 3+ | Total |
|---|
| Negative | 5 | 6 | 13 | 7 | 31 |
| 1+ | 15 | 1 | 11 | 3 | 30 |
| 2+ | 16 | 10 | 3 | 6 | 35 |
| 3+ | 11 | 2 | 0 | 1 | 14 |
| Total | 47 | 19 | 27 | 17 | 110 |
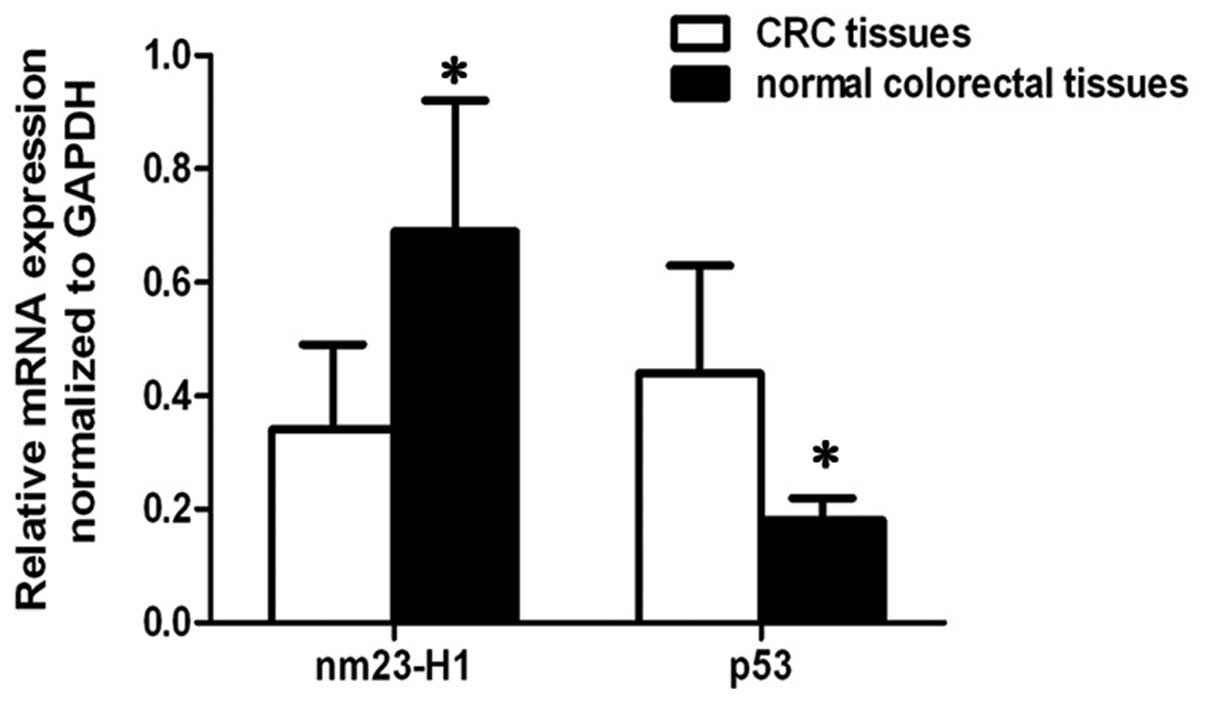
mRNA expression of Nm23-H1 and p53 in
CRC and normal colon tissues
As shown in Fig. 2,
the Nm23-H1 gene expression levels at the transcriptional level
were found to be significantly increased in the normal colorectal
tissues, compared with the CRC tissues (P<0.05). By contrast,
the mRNA expression levels of p53 were significantly decreased in
the normal colorectal tissues, compared with the CRC tissues
(P<0.05).
Association between the protein
expression levels of Nm23-H1 and p53 and prognosis
The overall survival (OS) rates of the 110 patients
with CRC were 4–102 months, with a median OS (mOS) rate of 55
months. The disease-free survival (DFS) rates of patients was 4–89
months, with a median DFS (mDFS) rate of 48 months. The OS and DFS
curves are presented in Fig. 3A and
B. In addition, further stratified analyses of prognosis with
different protein expression levels of Nm23-H1 and p53 was
performed. There was a statistical difference in the five-year
survival rate of patients with various Nm23-H1 and p53 expression
statuses (χ2=33.429, P<0.001). Patients with
Nm23-H1(+)/p53(−) tumors had improved prognosis, and the five-year
survival rate (83.8%) was significantly higher, compared with that
in patients with Nm23-H1(−)/p53(+) tumors (20.00%; P=0.008),
Nm23-H1(+)/p53(+) tumors (23.8%; P<0.001) and Nm23-H1(−)/p53(−)
tumors (30.8%; P<0.001), respectively. In addition, no
significant differences in prognosis were found among the latter
three subgroups (P>0.0125). The five-year DFS rate revealed a
similar trend as the five-year OS rate (χ2=18.108,
P<0.001). The five-year DFS rates of patients with
Nm23-H1(+)/p53(−) tumors (70.2%) were significantly higher,
compared with those in patients with Nm23-H1(+)/p53(+) tumors
(16.7%; P<0.001) and patients with Nm23-H1(−)/p53(−) tumors
(30.8%; P=0.002), however, no significant difference in five-year
DFS rates were found between patients with Nm23-H1(+)/p53(−) tumors
and patients with Nm23(−)/p53(+) tumors (20.00%; P=0.047; Table III). Taken together, these data
showed that patients with Nm23-H1(+)/p53(−) CRC tumors had improved
long-term survival rates (Fig.
3C-D).
 | Table III.Association between the protein
expression levels of Nm23-H1 and p53 and prognosis. |
Table III.
Association between the protein
expression levels of Nm23-H1 and p53 and prognosis.
|
| Number of
cases |
| Number of
cases |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Tumor | <5 years | ≥5 years | 5 y-OS (%) | <5 years | ≥5 years | 5 y-DFS (%) | Total |
|---|
|
Nm23-H1(−)/p53(+) | 4 | 1 | 20.0 | 4 | 1 | 20.0 | 5 |
|
Nm23-H1(+)/p53(+) | 32 | 10 | 23.8 | 35 | 7 | 16.7 | 42 |
|
Nm23-H1(−)/p53(−) | 18 | 8 | 30.8 | 18 | 8 | 30.8 | 26 |
|
Nm23-H1(+)/p53(−) | 6 | 31 | 83.8 | 11 | 26 | 70.2 | 37 |
| Total | 60 | 50 |
| 68 | 42 |
| 110 |
Discussion
Molecular tumor markers hold considerable promise
for accurately predicting the recurrence and progression of cancer
in patients. At present, several markers have been reported to
predict the prognosis of patients with CRC, including p53, ki67 and
matrix metalloproteinase-2 (18). The
Nm23 gene is located at the long arm of chromosome 17 and consists
of three subtypes, namely H1, H2 and H3 (23). Nm23-H1 is a tumor metastasis
suppressor gene, and numerous studies have shown that it is crucial
in regulating cell proliferation and differentiation (24,25). Of
note, Nm23-H1 has been shown to be negatively correlated with tumor
metastases and prognosis in oophoroma (26) and lung cancer (27). However, its involvement in CRC remains
to be fully elucidated (28,29). In the present study, Nm23-H1 protein
was positively expressed in 63 of 110 CRC tumor cases, whereas
almost all normal colorectal tissues were found to be
Nm23-H1-positive (48/53). The gene expression levels of Nm23-H1
were found to be significantly increased in normal colorectal
tissues, compared with CRC tissues. The protein expression of
Nm23-H1 further decreased in samples with lymph node metastases. It
has been revealed (30) that Nm23-H1
can inhibit metastasis in lung cancer by inhibiting
epithelial-mesenchymal transition, however, the mechanism of action
of Nm23-H1 in CRC remains to be elucidated. It is may be that the
high expression level of Nm23-H1 is involved in decreasing CRC cell
infiltration and metastasis by reducing tumor cell motility. In the
present study, significant differences in the expression and
pathological patterns of Nm23-H1 were found. The data showed that
the rates of positive protein expression of Nm23-H1 in the
undifferentiated carcinoma and mucinous carcinoma were lower,
compared with that of adenocarcinoma, which may be one of the
reasons for poor prognosis.
The p53 gene is a tumor suppressor gene located in
the short arm of chromosome 17, and is key in controlling tumor
evolution and progression (31). In
normal cells, p53 can induce cell cycle arrest at the G0/G1 phase
and cell apoptosis; and when it is mutated, as in the case of
several types of tumor, p53 loses its regulatory role and promotes
tumor progression (32). Deregulation
of cell-cycle machinery involving alterations in p53 is common in
bladder cancer (33), breast cancer
(34), lung cancer (35) and CRC (36). Significant differences in p53 staining
between CRC tissues and normal control tissues have been reported
in previous studies (17,18). The results of studies have varied;
certain studies have revealed that the protein expression of p53 is
lower in CRC tissues, compared with normal colorectal tissues
(37), whereas other studies have
shown its expression was higher in CRC (38,39).
Significant differences in the protein and mRNA expression levels
of p53 between CRC and normal colorectal tissues were also found in
the present study. The protein and mRNA expression levels of p53
were markedly higher in the CRC tissues, compared with the normal
controls tissues The cohort of patients the present study comprised
patients with stage II and III CRC. This may be one of the reasons
causing high protein expression levels of p53. In addition, other
studies have demonstrated that tumor differentiation is correlated
with the expression of p53 (35,36,38);
wherein, the lower the degree of tumor differentiation, the higher
the protein expression of p53. p53 has been confirmed as an
important prognostic marker for patients with CRC (28). However, in the present study, no
significant correlation was found between the protein expression of
p53 and the clinical pathological features of the patients with CRC
or their tumors.
As colorectal tumorigenesis is a complex process, it
is evident that the usefulness of a single marker for the
prediction of prognosis is limited. Therefore, identifying novel
molecular markers or the combined detection of two or more tumor
markers is warranted.
In the present study, significant differences were
found in the five-year OS rates and five-year DFS rates of patients
with CRC with different Nm23-H1 and p53 expression status. Patients
with Nm23-H1(+)/p53 (−) tumors had a good prognosis, and their
five-year OS and five-year DFS rates were markedly higher, compared
with those observed in patients with other Nm23-H1/p53 combined
protein expression statuses. The functional association between
Nm23-H1 and p53 proteins remains to be elucidated. The regulation
of Nm23-H1 by p53 differs in different types of cancer. For
example, a negative correlation has been reported in gastric cancer
(40). Other studies have indicated
marked heterogeneity in the expression of p53 and Nm23-H1 in
squamous cell lung cancer (41). In
CRC, the expression of Nm23-H1 enhances the inhibition of tumor
metastasis and tumor cell invasion, whereas negative expression of
the p53 protein is associated with the inhibition of cancer cell
proliferation (42). It is possible
that these two proteins act synergistically and affect each other,
leading to decreases in tumor cell growth and the invasive ability
of cancer cells; thus, prolonging patient survival rates.
Therefore, the combined detection of the Nm23-H1 and p53 proteins
may provide clinically useful information on the biological
behavior and prognosis of CRC. Based on the results of the present
study, it can be concluded that the combined detection of Nm23-H1
and p53 offers potential for predicting the prognosis of patients
with CRC with stage II and III tumors. However, future
investigations clarifying the functional association of these two
mediators in the progression of CRC and other tumors are
warranted.
It can be concluded that two different protein
markers be considered when evaluating the clinical outcome of
patients with CRC. The combined detection of the protein expression
of Nm23-H1 and p53 may provide an index for accurately predicting
the prognosis of CRC by providing information on malignancy and
biological behaviors.
Acknowledgements
This study was supported by funds from the National
Natural Science Foundation of China (grant no. 81172169), the Key
Technologies R & D Programs of Shaanxi province (grant nos.
2015SF041, 2015SF037 and S2013SF3846), the Fundamental Research
Funds for the Central University (grant no. 2015gjhz18) and the
Research Fund for the Doctoral Program of Higher Education of China
(grant no. 20130201120079).
References
|
1
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nie SF, Yao X, Zhu GB, Zhang JR, Xu YH and
Wang X: 1:2 Matched case-control study on risk factors of
colorectal cancer in Wuhan. China Public Health. 18:1482–1784.
2002.
|
|
3
|
Li J, Guo BC, Sun LR, Wang JW, Fu XH,
Zhang SZ, Poston G and Ding KF: TNM staging of colorectal cancer
should be reconsidered by T stage weighting. Worl J Gastroenterol.
20:5104–5112. 2014. View Article : Google Scholar
|
|
4
|
Uen YH, Lin SR, Wu DC, Su YC, Wu JY, Cheng
TL, Chi CW and Wang JY: Prognostic significance of multiple
molecular markers for patients with stage II colorectal cancer
undergoing curative resection. Ann Surg. 246:1040–1046. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Voorneveld PW, Jacobs RJ, Kodach LL and
Hardwick JC: A meta-analysis of SMAD4 immunohistochemistry as a
prognostic marker in colorectal cancer. Transl Oncol. 8:18–24.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim HO, Kim BG, Cha SJ, Park YG and Lee
TJ: Clinicopathologic significance of BRAF mutation and
extracellular signal regulated kinase 1/2 expression in patients
with a colorectal adenocarcinoma. Ann Coloproctol. 31:9–15. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu HW, Gao LD and Wei GH: hMSH2 and nm23
expression in sporadic colorectal cancer and its clinical
significance. Asian Pac J Cancer Prev. 14:1995–1998. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki E, Ota T, Tsukuda K, Okita A,
Matsuoka K, Murakami M, Doihara H and Shimizu N: Nm23-H1 reduces in
vitro cell migration and the liver metastatic potential of colon
cancer cells by regulating myosin light chain phosphorylation. Int
J Cancer. 108:207–211. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bal A, Joshi K, Logasundaram R, Radotra BD
and Singh R: Expression of nm23 in the spectrum of pre-invasive,
invasive and metastatic breast lesions. Diagn Pathol. 3:232008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakayama T, Ohtsuru A, Nakao K, Shima M,
Nakata K, Watanabe K, Ishii N, Kimura N and Nagataki S: Expression
in human hepatocellular carcinoma of nucleoside diphosphate kinase,
a homologue of the nm23 gene product. J Natl Cancer Inst.
84:1349–1354. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Z, Lu P, Zhang H, Xu H, Gao N, Li M
and Liu C: Nestin positively regulates the Wnt/ß-catenin pathway
and the proliferation, survival and invasiveness of breast cancer
stem cells. Breast Cancer Res. 16:4082014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mandai M, Konishi I, Koshiyama M, Mori T,
Arao S, Tashiro H, Okamura H, Normura H, Hiai H and Fukumoto M:
Expression of metastasis-related nm23-H1 and nm23-H2 genes in
ovarian carcinomas: Correlation with clinicopathology, EGFR,
c-erbB-2, and c-erbB-3 genes, and sex steroid receptor expression.
Cancer Res. 54:1825–1830. 1994.PubMed/NCBI
|
|
13
|
Cheok CF, Verma CS, Baselqa J and Lane DP:
Translating p53 into clinic. Nat Rev Clin Oncol. 8:25–37. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vogelstein B, Lane DP and Levine AJ:
Suring the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu W, Yu YH, Ouyang XN, Wang L, Wu YM,
Chen J and Xiong XS: Clinical significance of p53 and Ki67
expression in gastric cancer. World J. Gastroenterol. 19:367–373.
2011.
|
|
16
|
Ma LL, Zhe W, Wang YQ, Cong XL and Zhang
XM: Expression and clinical significance of Survivin and p53 in
breast cancer. Chin J Exp Surg. 31:2839–2840. 2014.
|
|
17
|
Zhang YX and Wang Y: Clinical significance
and expression of p53 in colorectal cancer. World J Integrated
Traditional and Western Med. 2:294–296. 2007.
|
|
18
|
Xiao ZW, He N, Cao JQ, Li ZJB and Zhu ZM:
The relationship between the expression of MMP-2 and p53 and tumor
invasion and metastasis in colorectal carcinoma. The Practical J
Cancer. 24:39–43. 2009.
|
|
19
|
Yin H, Xu L and Yao HW: New opinions of
colorectal cancer in 2010. Chin J Practical Surg. 30:764–768.
2010.
|
|
20
|
Hyslop T, Weinberg DS, Schulz S, Barkun A
and Waldman SA: Analytic lymph node number establishes staging
accuracy by occult tumor burden in colorectal cancer. J Surg Oncol.
106:24–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao P, Song YX, Wang ZN, Xu YY, Tong LL,
Sun JX, Yu M and Xu HM: Is the prediction of prognosis not improved
by the seventh edition of TNM classification for colorectal cancer?
Analysis of the surveillance, epidemiology, and end results (SEER)
database. BMC Cancer. 13:1232013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Backer JM, Mendola CE, Kovesdi I,
Fairhurst JL, O'Hara B, Eddy RL Jr, Shows TB, Mathew S, Murty VV
and Chaganti RS: Chromosomal localization and nucleoside
diphosphate kinase activity of human metastasis suppressor genes
nm23-1 and nm23-2. Oncogene. 8:497–502. 1993.PubMed/NCBI
|
|
24
|
Kim HD, Youn B, Kim TS, Kim SH, Shin HS
and Kim J: Regulators affecting the metastasis suppressor activity
of Nm23-H1. Mol Cell Biochem. 329:167–173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lombardi D, Lacombe ML and Paggi MG: Nm23:
Unraveling its biological function in cell differentiation. J Cell
Physiol. 182:144–149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang YQ and Li JC: Correlation of genetic
instablity of nm23H1 and clinicopathologic features of epithelial
in ovarian carcinoma. Ai Zheng. 25:713–717. 2006.(In Chinese).
PubMed/NCBI
|
|
27
|
Zheng HX, Cui YX, Shen DL, Peng A and He
YL: Influence of nm23-H1 gene transfection on expression of MMP-9
and TIMP-1 in lung cancer cells. Chin Clin Oncol. 20:104–107.
2015.
|
|
28
|
Dusonchet L, Corsale S, Migliavacca M,
Calò V, Bazan V, Amato A, Cammareri P, Totaro MS, Agnese V, Cascio
S, et al: Nm23-H1 expression does not predict clinical survival in
colorectal cancer patients. Oncol Rep. 10:1257–1263.
2003.PubMed/NCBI
|
|
29
|
Song JM, Kelton G and Wang JJ: Review
colorectal cancer screening: Current available methods. Chin
General Practice. 11:1115–1120. 2008.
|
|
30
|
Zhao R, Gong L, Li L, Guo L, Zhu D, Wu Z
and Zhou Q: Nm23-H1 is a negative regulator of TGF-β1-dependent
induction of epithelial-mesenchymal transition. Exp Cell Res.
319:740–749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Selivanova G: Wild type p53 reaction: From
lab bench to clinic. FEBS Lett. 588:2628–2638. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang YS, Graves B, Guerlavais V, Tovar C,
Packman K, To KH, Olson KA, Kesavan K, Gangurde P, Muknherjee A, et
al: Stapled α-helical peptide drug development: A potent dual
inhibitor of MDM2 and MDMX for p53-dependent canceer therapy. Proc
Natl Acad Sci USA. 110:E3445–E3454. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou XF, Zhang G and Tian Y: P53 status
correlates with the risk of recurrence in non-muscle invasive
bladder cancers treated with bacillus calmette Guérin: A
meta-analysis. PLoS One. 10:e01194762015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dolicanin Z, Velicković LJ, Djordjević B,
Visnjić M, Pesić I, Ristić A and Marianović V: Expression of
regulatory proteins and proliferative activity in relation to
phenotypic characteristics of upper urothelial carcinoma.
Vojnosanit Pregl. 68:567–574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
da Silva GN, Evangelista AF, Magalhães DA,
Macedo C, Búfalo MC, Sakamoto-Hojo ET, Passos GA and Saladori DM:
Expression of genes related to apoptosis, cell cycle and signaling
pathways are independent of TP53 status in urinary bladder cancer
cells. Mol Biol Rep. 38:4159–4170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen XY, Shi ZL and Li YH: Co-expression
of P-gp, GST, TOP II and p53 in colorectal carcinoma. J Clin Exp
Pathol. 18:608–610. 2002.
|
|
37
|
Lu Y, Gao J and Lu Y: Down-expression
pattern of Ku70 and p53 coexisted in colorectal cancer. Med Oncol.
32:982015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ye B, Wang XC, Lei L and You LS:
Expression and clinical significances of p53 and Ki67 in colorectal
carcinoma. The Practical J Cancer. 28:42–44. 2013.
|
|
39
|
Liu BW, Liu Y, Liu JR, Feng ZX and Liu T:
Prognositic effect of p53 expression in patients with completely
resected colorectal cancer. Tumour Biol. 35:9893–9896. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Geng QQ, Li Y, Tang CH, Li EX, Wu YY and
Zhang GJ: Expression and clinical significance of vascular
endothelial growth factor-C and nm23-H1 in stage II and III
colorectal carcinomas. Zhonghua Zhong Liu Za Zhi. 35:439–444.
2013.(In Chinese). PubMed/NCBI
|
|
41
|
Radović S, Dorić M, Hukić A, Babić M,
Kuskunović S and Spahović N: Immunohistochemical expression and
significance of Nm23 suppressor protein in primary gastric
adenocarcinoma. Bosn J Basic Med Sci. 13:72–77. 2013.PubMed/NCBI
|
|
42
|
Porebska I, Kosacka M, Wyrodek E and
Jankowska R: Expression of p53, bcl-2 and nm23 protein in squamous
cell lung cancer. Pneumonol Alergol Pol. 77:131–137. 2009.(In
Polish). PubMed/NCBI
|