Introduction
A growing body of evidence suggests that patients
with cancer develop antibodies against a variety of
tumor-associated antigens (TAA) (1–9). This has
led to the theory that autoantibodies may be used as a tool for the
early diagnosis of cancer. Natural immunoglobulin (Ig)M antibodies
have been associated with the recognition and elimination of
cancerous and precancerous cells (10,11). The
majority of research has focused on establishing cancer biomarkers
using IgG for the diagnosis or treatment of breast cancer, while
IgM autoantibodies have been insufficiently studied despite their
relevance in the early recognition of tumor antigens (12,13).
Strains of mice have been demonstrated to have different
susceptibilities to spontaneous breast cancer, DBA/2J being one of
the most susceptible, C57BL/6J one of the most resistant and BALB/c
being moderately susceptible (14).
The aim of the present study was to analyze the
patterns of recognition of 4T1 cell antigens using natural IgM from
the sera of mice with different levels of susceptibility to
spontaneous cancer, and to determine if there is any difference in
tumor recognition patterns among the strains in order to deduce the
putative natural IgM-recognizable antigens characteristic of the
different levels of cancer susceptibility.
Materials and methods
Cell extracts
4T1 mouse tumor cells [ATCC® CRL-2539;
donated by Dr Karen Manucharyan from the Instituto de
Investigaciones Biomédicas, Universidad Nacional Autónoma de México
(UNAM), Ciudad de México, Mexico] was cultured in RPMI 1640 medium
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and 1%
streptomycin/penicillin/amphotericin mixture (Gibco; Thermo Fisher
Scientific, Inc.) in 25 cm2 culture dishes (Corning
Incorporated, Corning, NY, USA), and incubated at 37°C in an
atmosphere of 98% humidity and 5% CO2. Cultures at a
confluence of 70–90% were collected by scraping and protein
extracts were obtained for two-dimensional (2D) electrophoresis as
previously described (12,13).
Mice and serum samples
A total of 10 healthy females (8 weeks old) of each
BALB/c, C57BL/6, and DBA/2J mouse strains were kept in the animal
facilities at the Instituto de Investigaciones Biomédicas, UNAM,
under controlled conditions of temperature (22°C), a relative
humidity of 50–60% and 12 h dark-light cycles, with lights on
between 7:00 a.m. and 9.00 p.m. The mice had free access to food
and water ad libitum. The Ethics Committee of the Institute
of Biomedical Research, UNAM approved this protocol (permission no.
2015–175). The mice were tail-bled on one occasion. The blood was
incubated at 4°C for 30 min and centrifuged at 1,306 × g for
10 min to obtain the serum, which was stored at −80°C until
use.
Immunoblot analysis of 2D images
2D immunoblots and image analysis were performed as
previously described (12,13). Briefly, the 2D immunoblots were
digitalized on a HP Scanjet G4050 scanner with a resolution of 300
dpi in a TIFF file format. All 2D immunoblots were analyzed using
the same settings for brightness, contrast and color to minimize
bias. The images were transferred to Adobe Photoshop CS5 (Adobe
Systems Europe, Ltd., Maidenhead, UK) to match them according to
spots present on all 2D immunoblots. The TIFF images were converted
to.1sc format, as required for analysis in PDQuest™ 2-D Analysis
Software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Master
images were created from the duplicates of the 2D immunoblot
images. Numbers of spots and their coordinates were determined on
the 2D immunoblots.
Clustering
To analyze the patterns of the IgM antibodies, a
clustering algorithm was applied. The 2D immunoblot images were
analyzed as previously described (13). Each 2D immunoblot master was divided
into 10 columns (pHi) ×10 rows (molecular weight, kDa). In each
grid, matrices were established, assigning a score of 0 if there
was no spot in the cell and 1 if there were ≥1 spots. The matrix
was converted into a vector by placing the n-th row immediately
after its predecessor. Thus, instead of a 10×10 matrix, a vector
was generated with 100 places containing values of 0 and 1. This
vector was used as the input for a python script to perform
complete linkage clustering with the hcluster package 0.2.0
(15). For this analysis, the
city-block metric was chosen, in which the distance between two
points is the sum of the absolute differences of their Cartesian
coordinates. The resulting hierarchical clustering was presented as
a dendrogram.
Statistical analysis
A paired two sample t-test for means was used to
analyze the total number of spots, and was performed using
Microsoft Office Excel 2010 (Microsoft Corporation, Redmond, WA,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results and Discussion
When tested on the 4T1 cell antigens, the sera from
the mice in all three groups displayed extremely different IgM
reactivity patterns. The master images obtained from the
immunoblots subsequent to processing with the PDQuest program
exhibited large and notable disparities in antigen recognition
among the three strains (Fig. 1).
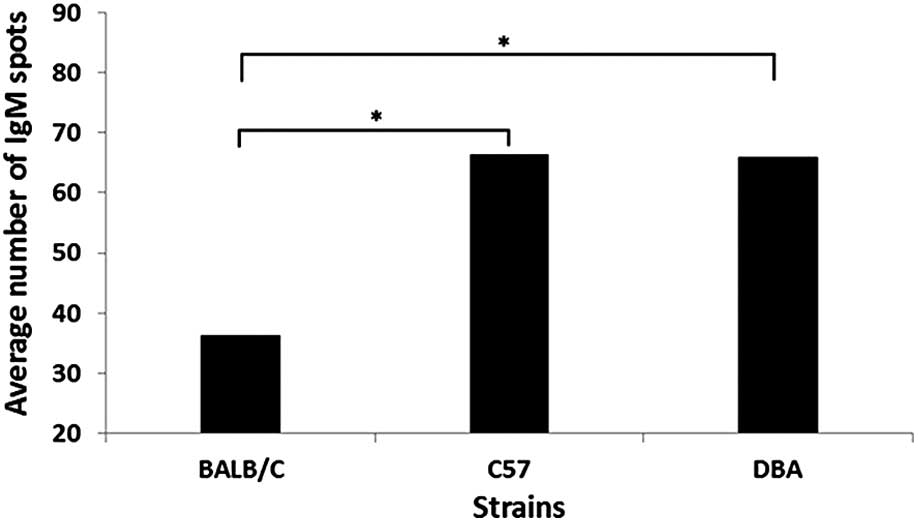
According to the average number of spots, no significant
differences in recognition spot numbers were observed among the
C57BL/6J and DBA/2J strains (P=0.9567). However, significant
differences in recognition spot numbers were observed among the
BALB/c and DBA/2J strains (P=0.017) and the BALB/c and C57BL/6J
strains (P=0.0053) (Fig. 2). This
indicates that the number of spots, reflecting the number of active
clones of IgM, is not decisive to determine susceptibility. In
addition, when the spots were analyzed according to their positions
on the blots, interspecies variations were much more evident than
individual intraspecies disparities.
The 2D immunoblots presented in Fig. 1 demonstrate notable differences in the
patterns of spot recognition between the DBA/2J and C57BL/6J
strains. The C57BL6 serum was markedly different from the DBA and
BALB/c sera, confirming the presence of strain-specific natural IgM
antibody repertoires. If the reactivity of the natural IgM
antibodies were merely neutral, it may be expected that the
different strain serum should exhibit a similar degree and scope of
IgM acuteness. When the 2D immunoblots were converted to digital
signatures and processed in order to group them according to
clustering algorithm (13), the
resulting dendrogram grouped the individuals almost perfectly
according to their respective strains (Fig. 3). This demonstrates that the method
perceived natural IgM antibody repertoire differences. With one
exception, all animals clustered according to their strain
genotype. All of them constituted homogeneous groups according to
their immunoreactivity. It appears that, rather than representing
random noise in the system, natural IgM antibodies are a repertoire
selected predominantly by the expression of a developmental genetic
program for V gene expression, without ligand-dependent selection
of clonal reactivities, which, according to Nobrega et al
(16), is designated as the
‘immunological homunculus’.
Notably, natural IgM antibody reactivities were
selectively directed towards a defined subset of all 4T1 antigens,
regardless of the amount of antigenic protein. This demonstrates
that the binding of natural IgM may be specific.
These results agree with those reported in (16), whereby different IgM
immunoreactivities of the BALB/c, DBA/2J and C57BL/6J strains
towards mouse liver extract antigens were observed. The results of
the current study reinforce the notion of the genetic control of,
in this case, the natural IgM antibody repertoire. It appears that
at birth there is a finite number of particular sets of
individually manifested IgMs that may be grouped according to
species-specific criteria; the natural IgM from mice of different
strains react differently to the presentation of an extract of
cancer cells (tumor antigens) to which they had not been previously
exposed. As DBA/2J, C57BL/6J and BALB/c mice have naturally
different susceptibilities to breast cancer (14), it may be assumed that IgM is able to
distinguish between antigens contributing in greater or lesser
measure to susceptibility to cancer.
The methodology used in the present study allowed
identification (by molecular weight and isoelectric point) of the
antigens that were more frequent, possibility indicating that they
are fundamental in the differences among the strain patterns
(Fig. 4; Table I). The identification of the
susceptibility-specific antigens is underway in future studies. The
determination of patterns of susceptibility to breast cancer in
raised mice is only a first step in defining the susceptibility
that humans present to cancer, and the question remains whether
patterns of natural IgM antibodies in humans may be used to
anticipate susceptibility to breast cancer. The possible functions
of natural IgM antibodies with generally greater or lesser
resistance in breast cancer require further investigation, but at
present their potential for immunodiagnosis is clear.
 | Table I.Frequency, isoelectric point and
molecular weight of the antigens marked in Fig. 4. |
Table I.
Frequency, isoelectric point and
molecular weight of the antigens marked in Fig. 4.
| Strain | Spot N | Frequency, % | ~Isoelectric
point | ~Molecular weight,
kDa |
|---|
| C57 | 1 | 90 | 4.05 | 73 |
|
| 2 | 100 | 7.1 | 103 |
|
| 3 | 100 | 6.9 | 69 |
|
| 4 | 90 | 7.8 | 13.5 |
| DAB | 1 | 90 | 5.2 | 52 |
|
| 2 | 80 | 5.3 | 68 |
|
| 3 | 80 | 5.6 | 53 |
In conclusion, the results of the current study
demonstrate that it is possible to segregate the IgM humoral immune
response toward cancer antigens according to the genetic background
of individuals. In addition, it is possible to identify the
recognized antigens that allow grouping or discriminate between the
different IgM antibodies expressed. The possible association of a
particular antigen with the susceptibility to cancer requires
further study, but the methodology applied in the present study may
allow the unveiling of candidates for this possible
association.
Acknowledgements
The current study received financial support from
the Consejo Nacional de Ciencia y Tecnología Mexico, Ciudad de
México, México (grant no. 151747) and the Programa de Apoyo a
Proyectos de Investigación e Innovación Tecnológica (PAPITT),
Dirección General de Asuntos del Personal Académico (DGAPA),
Universidad Nacional Autónoma de Mexico, Ciudad de México, México
(grant no. IN201715).
References
|
1
|
Tabernero MD, Lv LL and Anderson KS:
Autoantibody profiles as biomarkers of breast cancer. Cancer
Biomark. 6:247–256. 2010.PubMed/NCBI
|
|
2
|
Tan HT, Low J, Lim SG and Chung MC: Serum
autoantibodies as biomarkers for early cancer detection. FEBS J.
276:6880–6904. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Desmetz C, Bascoul-Mollevi C, Rochaix P,
Lamy PJ, Kramar A, Rouanet P, Maudelonde T, Mangé A and Solassol J:
Identification of a new panel of serum autoantibodies associated
with the presence of in situ carcinoma of the breast in younger
women. Clin Cancer Res. 15:4733–4741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Anderson KS, Ramachandran N, Wong J,
Raphael JV, Hainsworth E, Demirkan G, Cramer D, Aronzon D, Hodi FS,
Harris L, et al: Application of protein microarrays for multiplexed
detection of antibodies to tumor antigens in breast cancer. J
Proteome Res. 7:1490–1499. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan EM and Zhang J: Autoantibodies to
tumor-associated antigens: Reporters from the immune system.
Immunol Rev. 222:328–340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu H, Goodell V and Disis ML: Humoral
immunity directed against tumor-associated-antigens as potential
biomarkers for the early diagnosis of cancer. J Proteome Res.
7:1388–1394. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Finn OJ: Immune response as a biomarker
for cancer detection and a lot more. N Engl J Med. 353:1288–1290.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Storr SJ, Chakrabarti J, Barnes A, Murray
A, Chapman CJ and Robertson JF: Use of autoantibodies in breast
cancer screening and diagnosis. Expert Rev Anticancer Ther.
6:1215–1223. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anderson KS and LaBaer J: The sentinel
within: Exploiting the immune system for cancer biomarkers. J
Proteome Res. 4:1123–1133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vollmers HP and Brändlein S: Natural
antibodies and cancer. N Biotechnol. 25:294–298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Díaz-Zaragoza M, Hernández-Ávila R,
Viedma-Rodríguez R, Arenas-Aranda D and Ostoa-Saloma P: Natural and
adaptive IgM antibodies in the recognition of tumor-associated
antigens of breast cancer (Review). Oncol Rep. 34:1106–1114.
2015.PubMed/NCBI
|
|
12
|
Díaz-Zaragoza M, Hernández R and
Ostoa-Saloma P: 2D immunoblots show differential response of mouse
IgG and IgM antibodies to antigens of mammary carcinoma 4 T1 cells.
Cancer Cell Int. 14:92014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Díaz-Zaragoza M, Hernández-Ávila R,
Govezensky T, Mendoza L, Meneses-Ruíz DM and Ostoa-Saloma P:
Comparison patterns of 4 T1 antigens recognized by humoral immune
response mediated by IgG and IgM antibodies in female and male mice
with breast cancer using 2D-immnunoblots. Immunobiology.
220:1050–1058. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoag WG: Spontaneous cancer in mice. Ann N
Y Acad Sci. 108:805–831. 1963. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eads D: hcluster 0.2.0: A hierarcial
clustering package for Scipy. https://pypi.python.org/pypi/hcluster/0.2.0Accessed
March 4, 2016.
|
|
16
|
Nobrega A, Haury M, Grandien A, Malanchère
E, Sundblad A and Coutinho A: Global analysis of antibody
repertoires. II. Evidence for specificity, self-selection and the
immunological ‘homunculus’ of antibodies in normal serum. Eur J
Immunol. 23:2851–2859. 1993. View Article : Google Scholar : PubMed/NCBI
|