Introduction
Oral cancer is the most frequently observed cancer
of the head and neck region worldwide, with ~363,000 new cases
reported annually and a mortality rate of ~50% (1,2). The
tongue is a vital organ that serves an essential role in speech and
swallowing. Tongue cancer, a form of oral cancer, has become one of
the greatest challenges in the head and neck cancer field (3). It has been reported that tongue cancer
comprises between 22 and 49% of all oral cancer (4). Tongue cancer begins as a small lump and
may spread throughout the tongue and to the gums (5). It has been estimated that 6–7% of tongue
cancer occurs in patients <40 years old (6).
Tongue cancer may be caused by numerous factors,
including old age, geographical location, family history,
nutritional deficiencies, infectious agents, and chronic alcohol
and tobacco use (7), however, the
exact cause is unknown. A previous study demonstrated that cyclin
D1 was overexpressed in patients with anterior tongue cancer with
no history of tobacco and alcohol use, and postulated that it may
contribute to the development of this cancer. A total of 18 DEGs
were identified in this study (8).
Another study reported that tumor protein p53, BCL2 associated X
apoptosis regulator and BCL2 apoptosis regulator were associated
with squamous cell cancer of the tongue (9). Therefore, although previous studies have
identified a number of genes and proteins associated with tongue
cancer, the exact pathogenesis of the disease remains unknown.
The present study investigated gene expression
profiles to identify differentially-expressed genes (DEGs) between
tongue cancer from patients with a history of tobacco and/or
alcohol use (habit group) and tongue cancer from
non-habit-associated patients (non-habit group). Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analysis were then performed to analyze the DEGs.
Several key genes associated with habit-associated tongue cancer
were identified through protein-protein interaction (PPI) network
and functional module analysis. These results provide insight into
the molecular mechanisms underlying habit-associated tongue cancer.
In addition, the key DEGs identified are potential therapeutic
targets for the treatment of tongue cancer in patients with a
history of tobacco and/or alcohol use.
Materials and methods
Microarray data
The gene expression profile of microarray dataset
GSE42023 was obtained from the Gene Expression Omnibus (GEO)
database (www.ncbi.nlm.nih.gov/geo). This dataset was originally
produced using the HumanHT-8 v3.0 Gene Expression BeadChip Array
(Illumina, Inc., San Diego, CA, USA) (8). Gene expression data from 22 human
anterior tongue cancer tissue samples were analyzed in this study,
including 10 habit-associated samples, which were obtained from
patients who had a long history (>10 years) of tobacco and/or
alcohol use, in addition to 12 non-habit associated samples, which
were taken from patients who had no prior history of tobacco and/or
alcohol use. Details of the patients included in this dataset are
listed in Table I. The gene
expression data of all samples was pre-processed through background
correction, quantile normalization, probe summarization and probe
ID to gene symbol using the Robust Multi-array Average algorithm
(10) in the affy software package
(version 1.8.31) of Bioconductor (http://www.bioconductor.org/packages/release/bioc/html).
 | Table I.Details of patients with habit and
non-habit associated tongue cancer in the GSE42023 microarray
dataset. |
Table I.
Details of patients with habit and
non-habit associated tongue cancer in the GSE42023 microarray
dataset.
| Category | Sample number | Gender (M/F) | Age (years) | Tumor grade |
|---|
| Habit-associated
tongue cancer samples | 1 | M | 37 |
Moderately-differentiated |
|
| 2 | M | 45 |
Moderately-differentiated |
|
| 3 | M | 52 |
Moderately-differentiated |
|
| 4 | M | 42 |
Moderately-differentiated |
|
| 5 | M | 45 |
Well-differentiated |
|
| 6 | M | 42 |
Well-differentiated |
|
| 7 | M | 41 |
Well-differentiated |
|
| 8 | M | 52 |
Well-differentiated |
|
| 9 | M | 67 |
Well-differentiated |
|
| 10 | F | 48 |
Moderately-differentiated |
|
Non-habit-associated tongue cancer
samples | 1 | M | 30 |
Moderately-differentiated |
|
| 2 | M | 36 |
Moderately-differentiated |
|
| 3 | M | 37 |
Well-differentiated |
|
| 4 | F | 50 |
Well-differentiated |
|
| 5 | F | 80 |
Well-differentiated |
|
| 6 | F | 40 |
Moderately-differentiated |
|
| 7 | F | 25 |
Well-differentiated |
|
| 8 | F | 46 |
Moderately-differentiated |
|
| 9 | F | 50 |
Moderately-differentiated |
|
| 10 | F | 63 |
Well-differentiated |
|
| 11 | F | 70 |
Well-differentiated |
|
| 12 | F | 56 |
Well-differentiated |
DEG analysis
The Linear Models for Microarray Data software
package (version 3.16.8) (11) from
Bioconductor (version 2.12; http://www.bioconductor.org/packages/release/bioc/html/limma.html)
was used to identify DEGs between the habit and non-habit groups.
DEGs with a cutoff criteria of P<0.05 and |log2
fold-change| value ≥1 were used for screening.
GO and KEGG pathway enrichment
analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; version 6.7; https://david.ncifcrf.gov) (12) was used to identify the GO (13) biological process associated with the
DEGs identified. KEGG (14) pathway
enrichment analysis was subsequently used to identify the primary
signaling pathways the DEGs functioned in. P<0.05 calculated by
Fisher's exact test was used as the cutoff criterion for
statistically significant GO and KEGG enrichment analysis.
Screening for transcription factors
(TFs) and tumor-associated genes (TAGs)
TFs and TAGs were identified from the DEGs using the
Encyclopedia of DNA Elements database (https://www.encodeproject.org) (15) and the TAG database (http://www.binfo.ncku.edu.tw/TAG) (16), respectively.
PPI network construction
The Search Tool for the Retrieval of Interacting
Genes (STRING; version 9.05; http://string-db.org) database, which provides
experimental and predicted PPI information (17), was used to analyze the PPI network for
the DEGs. A confidence score >0.4 was chosen as the threshold
for a significant interaction. Finally, the PPI network for the
remaining DEGs was visualized using Cytoscape software (version
3.0.0; www.cytoscape.org) (18).
Screening and analysis of the
functional module
The BioNet Package (version 1.8.0; http://bionet.bioapps.biozentrum.uni-wuerzburg.de)
provides a set of statistics for the analysis of gene expression
data and biological networks (19).
The functional module for DEGs was obtained based on BioNet
analysis of the PPI network. A false discovery rate <0.005 was
used as the cutoff criterion for functional module screening. GO
and KEGG enrichment analysis of functional modules was performed
using DAVID, with a statistically significant cutoff criterion of
P<0.05.
Results
Identification of DEGs
The microarray dataset GSE42023 was obtained from
the GEO database in order to identify the DEGs between the habit
and non-habits groups. In total, 642 DEGs were identified in the
habit group compared with the non-habit group, including 200
upregulated and 442 downregulated DEGs.
GO and KEGG functional enrichment
analysis
GO enrichment analysis demonstrated that the
upregulated DEGs were enriched in 29 biological processes,
including regulation of apoptosis (P=0.00531), skeletal muscle
tissue development (P=0.00773) and positive regulation of nuclear
factor-kB transcription factor activity (P=0.00485) (Table II). The downregulated DEGs were
identified to be enriched in 39 biological processes, including fat
cell differentiation (P=0.00074), response to ultraviolet light
(P=0.00118) and embryonic pattern specification (P=0.01686)
(Table II).
 | Table II.Top 10 enriched GO functions for
upregulated and downregulated DEGs. |
Table II.
Top 10 enriched GO functions for
upregulated and downregulated DEGs.
| Type of DEG | GO no. | GO function | No. of DEGs
enriched |
P-valuea | Genes |
|---|
| Upregulated | GO:0042981 | Regulation of
apoptotic process | 21 | 0.00531 | TEX11,
MST4, FGF8, FHL2, RARG, DNAJC5,
APAF1, PRKCZ, TNFRSF10B, IFI27,
INSL3, CTH, PSMB10, MLLT11,
KDM2B, DAPK3, SCG2, MTDH, CLU,
SHQ1, GRM4 |
|
| GO:0007519 | Skeletal muscle
tissue development | 7 | 0.00773 | RCAN1,
ERBB2, TCF21, CHRNA1, ADAM12,
MYEF2, EP300 |
|
| GO:0051092 | Positive regulation
of NF-kB transcription factor activity | 5 | 0.00485 | TLR9,
PRKCZ, CTH, MTDH, CLU |
|
| GO:0021532 | Neural tube
patterning | 3 | 0.00499 | FGF8,
FOXA2, KDM2B |
|
| GO:0060425 | Lung
morphogenesis | 3 | 0.01026 | FGF8,
FOXA2, TCF21 |
|
| GO:0060337 | Type I
interferon-mediated signaling pathway | 3 | 0.04164 | OAS2,
IFI27, IFNA13 |
|
|
GO:0045945 | Positive regulation
of transcription from RNA polymerase III promoter | 2 | 0.00212 | ERBB2,
FOXA2 |
|
| GO:0006853 | Carnitine
shuttle | 2 | 0.00358 | CPT1A,
PRKAB2 |
|
| GO:0050860 | Negative regulation
of T cell receptor signaling pathway | 2 | 0.00643 | PTPN22,
ELF1 |
|
| GO:0071542 | Dopaminergic neuron
differentiation | 2 | 0.00755 | FGF8,
FOXA2 |
| Downregulated | GO:0045444 | Fat cell
differentiation | 11 | 0.00074 | CTBP1,
LRP6, RETN, WNT5B, TGFB1I1,
ALDH6A1, SLC2A4, AKT1, INHBB,
CREB5, CREBL2 |
|
| GO:0009411 | Response to UV | 9 | 0.00118 | TGFB1I1,
ALDH6A1, SLC2A4 |
|
| GO:0009880 | Embryonic pattern
specification | 5 | 0.01686 | AKT1,
INHBB, CREB5, CREBL2 |
|
| GO:0002089 | Lens morphogenesis
in camera-type eye | 4 | 0.00275 | MSH6,
ZRANB3, XPA, AKT1 |
|
| GO:0043044 | ATP-dependent
chromatin remodeling | 4 | 0.00985 | N4BP1,
PIK3R1, SPRTN, REV1, HYAL1 |
|
| GO:0035690 | Cellular response
to drug F | 4 | 0.02375 | LRP6,
TDGF1, TFAP2A, COBL, PCSK6 |
|
| GO:0071364 | Cellular response
to epidermal growth factor stimulus | 3 | 0.00459 | PVRL3,
TFAP2A, PROX1, CITED2 |
|
| GO:0010765 | Positive regulation
of sodium ion transport | 3 | 0.01115 | RBBP7,
RSF1, NASP, SMARCAD1 |
|
| GO:0021516 | Dorsal spinal cord
development | 3 | 0.01475 | CAD,
PPM1F, TXN, KCNH2 |
|
| GO:0086091 | Regulation of heart
rate by cardiac conduction | 3 | 0.01475 | CAD,
AKT1, TDGF1 |
KEGG enrichment analysis identified that the
upregulated DEGs were significantly enriched in 9 signaling
pathways, including that of calcium (P=0.01949), long-term
potentiation (P=0.00326) and the spliceosome (P=0.02543) (Table III). The downregulated DEGs were
significantly enriched in 5 signaling pathways, such as the
adipocytokine signaling pathway (P=0.00374), the B cell receptor
signaling pathway (P=0.00607) and the non-small cell lung
cancer-associated signaling pathway (P=0.03098) (Table III).
 | Table III.Enriched KEGG signaling pathways for
DEGs. |
Table III.
Enriched KEGG signaling pathways for
DEGs.
| Type of DEG | KEGG no. | KEGG signaling
pathway | No. of DEGs
enriched |
P-valuea | Genes |
|---|
| Upregulated | 4020 | Calcium signaling
pathway | 5 | 0.01949 | CHP2,
AVPR1B, CAMK2D, ERBB2, CALM2 |
|
| 4720 | Long-term
potentiation | 4 | 0.00326 | CHP2,
CAMK2D, EP300, CALM2 |
|
| 3040 | Spliceosome | 4 | 0.02543 | SNRPD1,
SRSF4, TXNL4A, CCDC12 |
|
| 4650 | Natural killer cell
mediated cytotoxicity | 4 | 0.03167 | RAET1E,
CHP2, TNFRSF10B, IFNA13 |
|
| 4012 | ErbB signaling
pathway | 3 | 0.04129 | CAMK2D,
ERBB2, GAB1 |
|
| 4210 | Apoptosis | 3 | 0.04129 | CHP2,
APAF1, TNFRSF10B |
|
| 511 | Other glycan
degradation | 2 | 0.00962 | GLB1,
GBA |
|
| 4744 |
Phototransduction | 2 | 0.02683 | GUCA1B,
CALM2 |
|
| 600 | Sphingolipid
metabolism | 2 | 0.04847 | GLB1,
GBA |
| Downregulated | 4920 | Adipocytokine
signaling pathway | 6 | 0.00374 | RXRB,
SLC2A4, LEPR, AKT1, CAMKK2,
MAPK10 |
|
| 4662 | B-cell receptor
signaling pathway | 6 | 0.00607 | RASGRP3,
PIK3AP1, AKT1, BTK, PIK3R1,
NFATC3 |
|
| 5223 | Non-small cell lung
cancer | 4 | 0.03098 | RXRB,
AKT1, PIK3R1, RASSF1 |
|
| 4012 | ErbB signaling
pathway | 5 | 0.04306 | AKT1,
EREG, PIK3R1, ELK1, MAPK10 |
|
| 5210 | Colorectal
cancer | 4 | 0.04781 | MSH6,
AKT1, PIK3R1, MAPK10 |
Screening for TFs and TAGs
A total of 31 DEGs were identified as TFs, including
10 upregulated DEGs [e.g., E1A binding protein p300 (EP300),
RARG and HOXB13] and 21 downregulated DEGs (e.g.
CTBP1, GTF2A1 and RORB) (Table IV). Furthermore, 41 DEGs were
identified as TAGs, including 13 upregulated DEGs [e.g., erb-b2
receptor tyrosine kinase 2 (ERBB2), FGF8 and
MLLT11] and 28 downregulated DEGs [e.g., AKT
serine/threonine kinase 1 (AKT1), MLF1 and
FGF20] (Table IV).
 | Table IV.TFs and TAGs among the DEGs. |
Table IV.
TFs and TAGs among the DEGs.
|
| TFs | TAGs |
|---|
|
|
|
|
|---|
| Type of DEG | No. of genes | Genes | No. of genes | Genes |
|---|
| Upregulated | 10 | EP300, RARG,
HOXB13, RORA, FOXA2, TCF21, ELF, SIX5, CCNT2, SUPT4H1 | 13 | ERBB2, FGF8,
MLLT11, MT1G, APAF1, HOXB13, TNFRSF10B, PDLIM4, FOXA2, DAPK3, CLU,
MTUS1, FHL2 |
| Downregulated | 21 | CTBP1, GTF2A1,
RORB, RXRB, HOXB4, TGFB1I1, LMX1B, NR2F6, KCNIP3, CDX4, SKIL,
GABPB2, MEIS3, NKX6-1, PAX3, ELK1, NFATC3, SIM1, FOXD1, PROX1,
RFX5 | 28 | MLF1, FGF20,
CTTN, SKIL, AKT1, EWSR1, ELK1, DCUN1D1, CTBP1, PTPN2, RBBP7,
FBXO32, TGFBI, SIRT3, NEO1, RARRES1, PER1, SPINK7, CBFA2T3, ENC1,
CREBL2, PROX1, RASSF1, MSH6, PAX3, DDX6, TFAP2A, DHX16 |
Construction of the PPI network
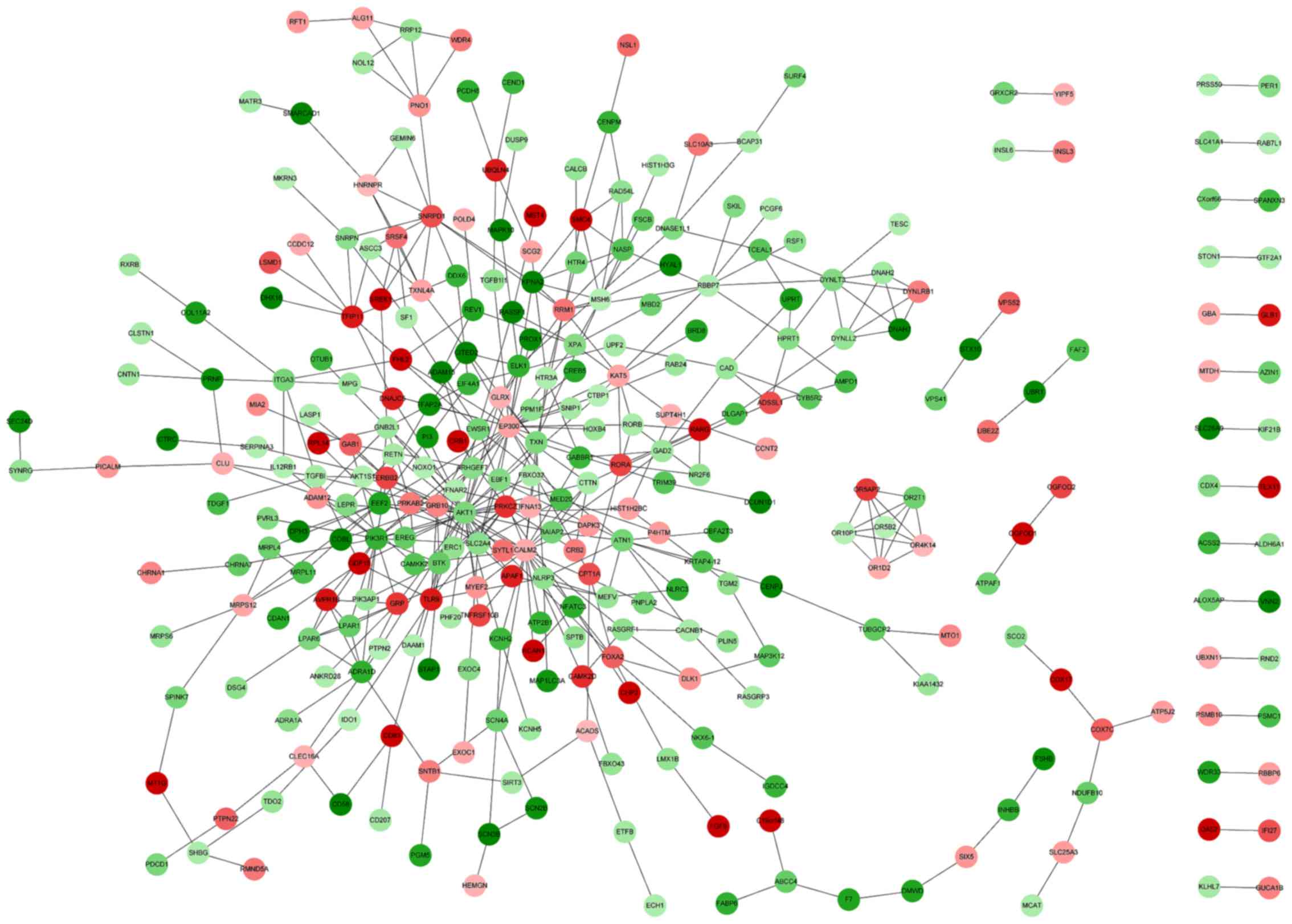
The DEG PPI network was constructed using STRING.
The resulting PPI network contained 330 nodes and 462 PPIs
(Fig. 1). The top 10% of nodes were
classified as having a high degree of connectivity in the PPI
network, these included AKT1, EP300, CALM2 and
PIK3R1 (Table V). AKT1
was identified to interact with ERBB2, epiregulin
(EREG) and EP300 (Fig.
1).
 | Table V.DEGs in the top 10% of nodes with a
high connectivity degree in the PPI. |
Table V.
DEGs in the top 10% of nodes with a
high connectivity degree in the PPI.
| DEG | STRING degree of
connectivity |
|---|
| AKT1 | 37 |
| EP300 | 25 |
| CALM2 | 23 |
| PIK3R1 | 18 |
| ATN1 | 11 |
| PRKCZ | 11 |
| SNRPD1 | 10 |
| EEF2 | 10 |
| MSH6 | 10 |
| ERBB2 | 9 |
| RBBP7 | 9 |
| GNB2L1 | 9 |
| TLR9 | 9 |
| RRM1 | 9 |
| BTK | 8 |
| SLC2A4 | 8 |
| TFIP11 | 8 |
| GRP | 8 |
| DYNLT3 | 8 |
| ADRA1D | 7 |
| NASP | 7 |
| ARHGEF7 | 7 |
| TXNL4A | 7 |
| TXN | 7 |
| GRB10 | 7 |
Construction and analysis of the
functional module
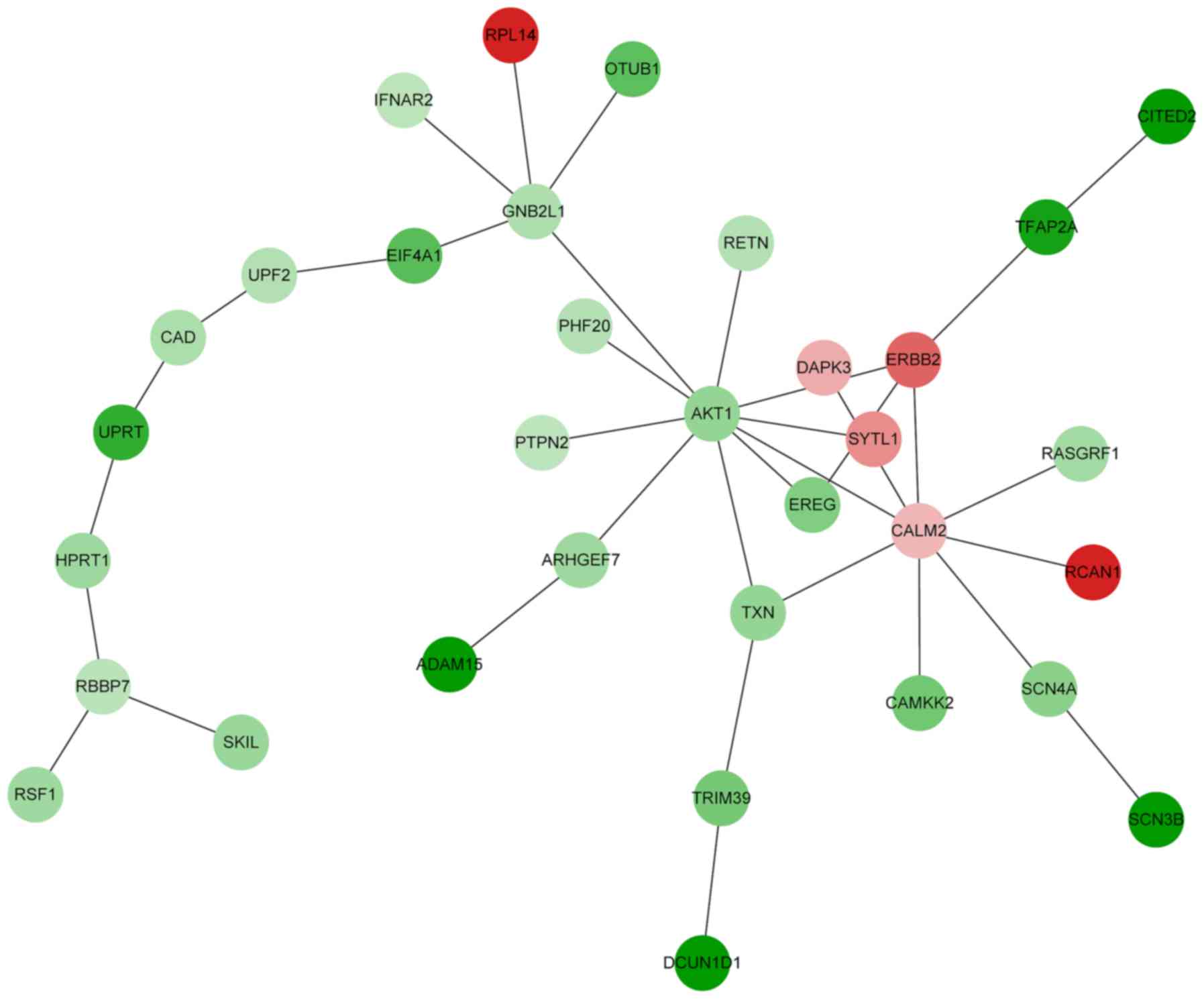
Based on the PPI network created, a functional
module was constructed by BioNet. The functional module contained
33 nodes and 35 PPIs (Fig. 2). The
connectivity degree of AKT1, CALM2, GNB2L1 and
ERBB2 was >4 in the functional module (data not shown).
DEGs in the functional module were enriched in 18 biological
processes defined by GO, including protein autophosphorylation
(P=0.0000425), female pregnancy (P=0.00034), positive regulation of
GTPase activity (P=0.00037) and cytokine-mediated signaling
(P=0.00586) (Table VI). KEGG
enrichment analysis demonstrated that the DEGs in the functional
module were enriched in 16 signaling pathways, such as the ErbB
signaling pathway (P=0.00126; e.g., EREG, ERBB2 and
AKT1), the focal adhesion pathway (P=0.01310; e.g.,
RASGRF1, ERBB2 and AKT1) and cancer-associated
pathways (P=0.04696; e.g., DAPK3, ERBB2 and
ATK1) (Table VII).
 | Table VI.Top 10 enriched GO functions for DEGs
in the functional module. |
Table VI.
Top 10 enriched GO functions for DEGs
in the functional module.
| GO no. | GO function | No. of DEGs
enriched |
P-valuea | Genes |
|---|
| GO:0046777 | Protein
autophosphorylation | 5 |
0.0000425 | CAD, DAPK3,
CAMKK2, ERBB2, AKT1 |
| GO:0007565 | Female
pregnancy | 4 | 0.00034 | UPRT, CAD,
CITED2, AKT1 |
| GO:0043547 | Positive regulation
of GTPase activity | 4 | 0.00037 | RASGRF1, ERBB2,
ARHGEF7, GNB2L1 |
| GO:0019221 | Cytokine-mediated
signaling pathway | 4 | 0.00586 | IFNAR2, EIF4A1,
EREG, PTPN2 |
| GO:0042059 | Negative regulation
of epidermal growth factor receptor signaling pathway | 3 |
0.0000918 | TFAP2A, PTPN2,
ARHGEF7 |
| GO:0042593 | Glucose
homeostasis | 3 | 0.00340 | PTPN2, TXN,
AKT1 |
| GO:0051151 | Negative regulation
of smooth muscle cell differentiation | 2 | 0.00016 | RCAN1,
EREG |
| GO:0006222 | UMP biosynthetic
process | 2 | 0.00025 | UPRT,
CAD |
| GO:0010765 | Positive regulation
of sodium ion transport | 2 | 0.00075 | AKT1,
SCN3B |
| GO:0021602 | Cranial nerve
morphogenesis | 2 | 0.00092 | TFAP2A,
CITED2 |
 | Table VII.Enriched KEGG signaling pathways for
DEGs in the functional module. |
Table VII.
Enriched KEGG signaling pathways for
DEGs in the functional module.
| KEGG no. | KEGG signaling
pathway | No. of DEGs
enriched |
P-valuea | Genes |
|---|
| 4012 | ErbB signaling
pathway | 3 | 0.00126 | EREG, ERBB2,
AKT1 |
| 4510 | Focal adhesion | 3 | 0.01310 | RASGRF1, ERBB2,
AKT1 |
| 5200 | Pathways in
cancer | 3 | 0.04696 | DAPK3, ERBB2,
AKT1 |
| 5219 | Bladder cancer | 2 | 0.00495 | DAPK3,
ERBB2 |
| 5213 | Endometrial
cancer | 2 | 0.00751 | ERBB2,
AKT1 |
| 5223 | Non-small cell lung
cancer | 2 | 0.00808 | ERBB2,
AKT1 |
| 5214 | Glioma | 2 | 0.01155 | AKT1,
CALM2 |
| 4920 | Adipocytokine
signaling pathway | 2 | 0.01260 | CAMKK2,
AKT1 |
| 5212 | Pancreatic
cancer | 2 | 0.01332 | ERBB2,
AKT1 |
| 5215 | Prostate
cancer | 2 | 0.02100 | ERBB2,
AKT1 |
Discussion
Tongue cancer has been associated with a number of
factors, including old age, geographical location and family
history (7). In addition, tongue
cancer is associated with certain habits, such as chewing betel
nuts, smoking and alcohol abuse (3).
Although knowledge of tongue cancer has progressed, the complex
pathogenesis of the cancer remains unclear. Therefore, there is a
requirement to investigate the molecular mechanisms underlying the
pathogenesis of tongue cancer and to screen for novel markers of
the disease. In the present study, the gene expression profiles of
habit- and non-habit-associated tongue cancer samples were analyzed
using bioinformatics methods. A total of 642 DEGs were identified
between the habit and non-habit groups. Through analysis of the
biological functions and pathways of the DEGs, a set of genes and
signaling pathways was identified to be associated with
habit-associated tongue cancer.
In the PPI network constructed, EP300 and
AKT1 exhibited a high degree of connectivity. EP300, also
known as p300, is a global transcriptional coactivator that
regulates the activity of numerous DNA-binding transcription
factors that are associated with a wide array of cellular
activities, such as cell growth and differentiation (20,21), which
are increased in uncontrolled malignant tumors (22). EP300 has been found to be involved in
DNA repair synthesis through its interaction with proliferating
cell nuclear antigen, which is essential for DNA replication
(23,24). To the best of our knowledge, there is
no evidence that EP300 is associated with habit-associated
tongue cancer currently. However, EP300 has been found to promote
cancer progression in the prostate (25) and colon (26). Thus, EP300 may serve a role in the
development of habit-associated tongue cancer, likely through
regulating cell growth.
AKT1 belongs to the Akt/protein kinase B subfamily
of serine/threonine kinases, which is frequently hyperactivated in
human cancer (27). The AKT family
(AKT1-3) has been found to integrate extracellular signals in
several cellular processes, including growth, proliferation,
differentiation, migration and survival (28). Numerous studies have demonstrated that
the phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of
rapamycin pathway serves an essential role in apoptosis and is
frequently activated in numerous types of human cancer, such as
head and neck squamous cell carcinoma (29,30),
prostate cancer (31), breast cancer
(32) and colorectal cancer (33). Cancer cells have a higher
proliferation rate compared with wild-type cells and frequently
lose the ability to undergo apoptosis (18). A previous study reported that
activated AKT regulates its downstream targets to increase
proliferation and decrease apoptosis in cells (34). AKT activation has been described as an
early cellular response to carcinogen exposure and may be a key
step in environmental carcinogenesis (35). In the current study, AKT1 was
identified to be significantly functionally enriched in
cancer-associated signaling pathways. The overexpression of AKT has
been detected in a variety of cancer types, including tongue cancer
(36), head and neck squamous cell
carcinoma (37), ovarian cancer
(38) and prostate cancer (39). There is no evidence, to the best of
our knowledge, that AKT1 is associated with habit-associated
tongue cancer at present. However, AKT1 may be associated with the
development of habit-associated tongue cancer via the regulation of
cell proliferation and differentiation.
In the current study, AKT1, ERBB2 and
EREG were demonstrated to be significantly functionally
enriched in the ErbB signaling pathway. The ErbB signaling pathway
regulates cell migration and invasion in normal and tumor mammary
epithelial cells (40). The ErbB
family, which consists of four members [epidermal growth factor
receptor (EGFR), ERBB2, ERBB3 and ERBB4], plays an important role
in cell proliferation and survival in numerous epithelial
malignancies (41). ERBB2 was
predicted to be a TAG in the current study. The overexpression of
ERBB2 particularly occurs with a high frequency in breast cancer
(42). In addition, Silva et
al (43) reported that ERBB2
expression is associated with the 10-year survival of patients with
tongue cancer (43), indicating that
ERBB2 serves an important role in tongue cancer development and
progression. However, to the best of our knowledge, there have been
no reports of an association between ERBB2 and habit-associated
tongue cancer thus far. EGFR regulates cell motility, invasion and
proliferation (44). EGFR
mutations have been identified to activate anti-apoptotic signaling
pathways, such as PI3K/AKT/mTOR and mitogen-activated protein
kinase (45). EREG, as a ligand of
EGFR, stimulates the EGFR signaling pathway, which promotes the
metastasis of breast cancer cells (46). The results of the current study
identified that AKT1 interacts with EREG in the PPI network. In
addition, AKT1 and ERBB2 were classified as oncogenes using the TAG
database. These results indicate that AKT1, ERBB2 and EREG are
associated with the tumorigenesis of habit-associated tongue cancer
and are potential therapeutic targets for the treatment of this
cancer.
The grade of the tumor samples in the microarray
dataset used in the present study was different between the habit
and non-habit groups (Table I). The
grade of the tumor may impact gene expression, so this should be
taken into consideration when interpreting the results. In future,
a more accurate comparison could be made if tumor samples of
different grades were divided into subgroups. In addition, the
results of the present study will be validated experimentally.
In conclusion, the present study identified key DEGs
in habit-associated tongue cancer. These DEGs, such as AKT1,
EP300, ERBB2 and EREG, may serve important
roles in the tumorigenesis of habit-associated tongue cancer and
could be used as therapeutic targets for the treatment of this
cancer. However, further experiments are required to verify the
results of the current study and increase our understanding of the
pathogenesis of habit-associated tongue cancer.
References
|
1
|
Böckelman C, Hagström J, Mäkinen L,
Keski-Säntti H, Häyry V, Lundin J, Atula T, Ristimäki A and Haglund
C: High CIP2A immunoreactivity is an independent prognostic
indicator in early-stage tongue cancer. Br J Cancer. 104:1890–1895.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sano D and Myers JN: Metastasis of
squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev.
26:645–662. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gosselin BJ: Malignant Tumors of the
Mobile Tongue. Medscape. Jun;2011.http://emedicine.medscape.com/article/847428-overviewAccessed
April 3, 2015.
|
|
4
|
Liang XH, Lewis J, Foote R, Smith D and
Kademani D: Prevalence and significance of human papillomavirus in
oral tongue cancer: The Mayo Clinic experience. J Oral Maxillofac
Surg. 66:1875–1880. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Z, Wang H and Li Q: Tongue tumor
detection in medical hyperspectral images. Sensors. 12:162–174.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garavello W, Spreafico R and Gaini RM:
Oral tongue cancer in young patients: A matched analysis. Oral
Oncol. 43:894–897. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moore SR, Johnson NW, Pierce AM and Wilson
DF: The epidemiology of tongue cancer: a review of global
incidence. Oral Dis. 6:75–84. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sebastian P, Babu JM, Prathibha R,
Hariharan R and Pillai MR: Anterior tongue cancer with no history
of tobacco and alcohol use may be a distinct molecular and clinical
entity. J Oral Pathol Med. 43:593–599. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie X, Clausen OP, De Angelis P and Boysen
M: The prognostic value of spontaneous apoptosis, Bax, Bcl-2, and
p53 in oral squamous cell carcinoma of the tongue. Cancer.
86:913–920. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seo J and Hoffman EP: Probe set
algorithms: Is there a rational best bet? BMC Bioinformatics.
7:3952006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wettenhall JM and Smyth GK: limmaGUI: A
graphical user interface for linear modeling of microarray data.
Bioinformatics. 20:3705–3706. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene Functional Classification Tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gene Ontology Consortium, . Gene ontology
consortium: Going forward. Nucleic Acids Res. 43:D1049–D1056. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
ENCODE Project Consortium, . An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen JS, Hung WS, Chan HH, Tsai SJ and Sun
HS: In silico identification of oncogenic potential of fyn-related
kinase in hepatocellular carcinoma. Bioinformatics. 29:420–427.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Beisser D, Klau GW, Dandekar T, Müller T
and Dittrich MT: BioNet An R-Package for the functional analysis of
biological networks. Bioinformatics. 26:1129–1130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogryzko VV, Schiltz RL, Russanova V,
Howard BH and Nakatani Y: The transcriptional coactivators p300 and
CBP are histone acetyltransferases. Cell. 87:953–959. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalkhoven E: CBP and p300: HATs for
different occasions. Biochem Pharmacol. 68:1145–1155. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lahtz C and Pfeifer GP: Epigenetic changes
of DNA repair genes in cancer. J Mol Cell Biol. 3:51–58. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hasan S, Hassa PO, Imhof R and Hottiger
MO: Transcription coactivator p300 binds PCNA and may have a role
in DNA repair synthesis. Nature. 410:387–391. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moldovan GL, Pfander B and Jentsch S:
PCNA, the maestro of the replication fork. Cell. 129:665–679. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Debes JD, Sebo TJ, Lohse CM, Murphy LM,
Haugen DA and Tindall DJ: p300 in prostate cancer proliferation and
progression. Cancer Res. 63:7638–7640. 2003.PubMed/NCBI
|
|
26
|
Ionov Y, Matsui S and Cowell JK: A role
for p300/CREB binding protein genes in promoting cancer progression
in colon cancer cell lines with microsatellite instability. Proc
Natl Acad Sci USA. 101:1273–1278. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ju X, Katiyar S, Wang C, Liu M, Jiao X, Li
S, Zhou J, Turner J, Lisanti MP, Russell RG, et al: Akt1 governs
breast cancer progression in vivo. Proc Natl Acad Sci USA.
104:7438–7443. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Freeman-Cook KD, Autry C, Borzillo G,
Gordon D, BarbacciTobin E, Bernardo V, Briere D, Clark T, Corbett
M, Jakubczak J, et al: Design of selective, ATP-competitive
inhibitors of Akt. J Med Chem. 53:4615–4622. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pfisterer K, Fusi A, Klinghammer K,
Knödler M, Nonnenmacher A and Keilholz U: PI3K/PTEN/AKT/mTOR
polymorphisms: association with clinical outcome in patients with
head and neck squamous cell carcinoma receiving cetuximab-docetax.
Head Neck. 37:471–478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Amornphimoltham P, Patel V, Molinolo A and
Gutkind JS: Head and Neck Cancer and the PI3K/Akt/mTOR Signaling
Network: Novel Molecular Targeted Therapies. Signaling Pathways in
Squamous Cancer. pp407–pp429. 2010.
|
|
31
|
Kong L, Schäfer G, Bu H, Zhang Y, Zhang Y
and Klocker H: Lamin A/C protein is overexpressed in
tissue-invading prostate cancer and promotes prostate cancer cell
growth, migration and invasion through the PI3K/AKT/PTEN pathway.
Carcinogenesis. 33:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Castaneda CA, CortesFunes H, Gomez HL and
Ciruelos EM: The phosphatidyl inositol 3-kinase/AKT signaling
pathway in breast cancer. Cancer Metastasis Rev. 29:751–759. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Eide PW, Cekaite L, Danielsen SA,
Eilertsen IA, Kjenseth A, Fykerud TA, Ågesen TH, Bruun J, Rivedal
E, Lothe RA and Leithe E: NEDD4 is overexpressed in colorectal
cancer and promotes colonic cell growth independently of the
PI3K/PTEN/AKT pathway. Cell Signal. 25:12–18. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weber SM, Bornstein S, Li Y, Malkoski SP,
Wang D, Rustgi AK, KuleszMartin MF, Wang XJ and Lu SL:
Tobacco-specific carcinogen nitrosamine
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone induces AKT
activation in head and neck epithelia. Int J Oncol. 39:1193–1198.
2011.PubMed/NCBI
|
|
35
|
West KA, Brognard J, Clark AS, Linnoila
IR, Yang X, Swain SM, Harris C, Belinsky S and Dennis PA: Rapid Akt
activation by nicotine and a tobacco carcinogen modulates the
phenotype of normal human airway epithelial cells. J Clin Invest.
111:81–90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Massarelli E, Liu DD, Lee JJ, ElNaggar AK,
Lo Muzio L, Staibano S, De Placido S, Myers JN and
Papadimitrakopoulou VA: Akt activation correlates with adverse
outcome in tongue cancer. Cancer. 104:2430–2436. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Amornphimoltham P, Sriuranpong V, Patel V,
Benavides F, Conti CJ, Sauk J, Sausville EA, Molinolo AA and
Gutkind JS: Persistent activation of the Akt pathway in head and
neck squamous cell carcinoma: A potential target for UCN-01. Clin
Cancer Res. 10:4029–4037. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Altomare DA, Wang HQ, Skele KL, De Rienzo
A, Klein-Szanto AJ, Godwin AK and Testa JR: AKT and mTOR
phosphorylation is frequently detected in ovarian cancer and can be
targeted to disrupt ovarian tumor cell growth. Oncogene.
23:5853–5857. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lundstrom TS and Sobel JD: Antibiotics for
gram-positive bacterial infections: Vancomycin,
quinupristin-dalfopristin, linezolid, and daptomycin. Infect Dis
Clin North Am. 18651–668. (x)2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hardy KM, Booth BW, Hendrix MJ, Salomon DS
and Strizzi L: ErbB/EGF signaling and EMT in mammary development
and breast cancer. J Mammary Gland Biol Neoplasia. 15:191–199.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sosa MS, LopezHaber C, Yang C, Wang H,
Lemmon MA, Busillo JM, Luo J, Benovic JL, KleinSzanto A, Yagi H, et
al: Identification of the Rac-GEF P-Rex1 as an essential mediator
of ErbB signaling in breast cancer. Mol Cell. 40:877–892. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Silva SD, Perez DE, Alves FA, Nishimoto
IN, Pinto CA, Kowalski LP and Graner E: ErbB2 and fatty acid
synthase (FAS) expression in 102 squamous cell carcinomas of the
tongue: Correlation with clinical outcomes. Oral Oncol. 44:484–490.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lu Z, Jiang G, BlumeJensen P and Hunter T:
Epidermal growth factor-induced tumor cell invasion and metastasis
initiated by dephosphorylation and downregulation of focal adhesion
kinase. Mol Cell Biol. 21:4016–4031. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Uribe P and Gonzalez S: Epidermal growth
factor receptor (EGFR) and squamous cell carcinoma of the skin:
Molecular bases for EGFR-targeted therapy. Pathol Res Pract.
207:337–342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nickerson NK, Gilmore JL, Allen KT, Riese
DJ II, Nephew KP and Foley J: EGFR-Ligand signaling in breast
cancer metastasis: Recurring developmental themes. 2011.
|