Introduction
Cancer has long been threatening human health; thus,
it is urgent to identify novel effective therapeutic agents and
methods for the treatment of cancer. Previous studies have
demonstrated that the extracts of madder, a type of traditional
Chinese medicine, may be associated with a reduced risk of certain
cancers (1). Among these extracts,
the anticancer activity of mollugin has been shown to be strong in
various types of human cancer cells (2).
Cancer cells produce the majority of their energy
via the glycolytic pathway under aerobic conditions (known as the
Warburg effect), which is a feature of the majority of cancer cells
(3,4).
The Warburg effect in cancer cells, caused by dysfunctional
oxidative phosphorylation, sustains glycolysis, which is
responsible for tumor initiation, progression and metastasis
(5,6).
Consequently, the molecular basis of aerobic glycolysis in cancer
has been biochemically investigated, and deregulated metabolism is
gaining recognition as a hallmark of cancer cells, and is being
explored for therapeutic potential.
It is widely accepted that specific expression of
pyruvate kinase isoenzyme M2 (PKM2) in cancer cells contributes to
this aerobic glycolysis phenotype (7). Pyruvate kinase converts
phosphoenolpyruvate into pyruvate, catalyzing the rate-limiting
step of glycolysis (8). The M1
isoenzyme of pyruvate kinase is present in adult tissues, whereas
PKM2 is a spliceosome variant detected in embryonic and cancer
cells. PKM2 expression in malignant cells is the result of the
tumor microenvironment, and is responsible for maintaining a
glycolytic phenotype (9). PKM2, which
has been identified as the predominant cause of the Warburg effect
in cancer cells, is essential in cancer metabolism and growth
(8,10).
The present study revealed that FFJ-5, a
characterized naphthoquinone modifier of mollugin, possessed
anticancer activities and induced cell apoptosis (2). It was observed that FFJ-5 inhibited
cancer cell proliferation, exerting its anticancer effects via
inhibiting the expression of PKM2.
Materials and methods
Antibodies, reagents and test
compounds
Rabbit monoclonal anti-PKM2 (1:1,000; catalog no.
4053), rabbit monoclonal anti-caspase-3 (1:1,000; catalog no.
9665), rabbit monoclonal anti-cleaved caspase-3 (Asp175; 1:1,000;
catalog no. 9664), rabbit monoclonal anti-poly (ADP-ribose)
polymerase (PARP) (1:1,000; catalog no. 9532), rabbit anti-human
cleaved PARP (Asp214; 1:1,000; catalog no. 5625), mouse monoclonal
anti-phosphorylated (p)-epidermal growth factor receptor (EGFR)
(p-Tyr1068-EGFR) (1:1,000; catalog no. 2236), rabbit monoclonal
anti-Akt (1:1,000; catalog no. 4685) and rabbit monoclonal
anti-p-Akt (p-Ser473-Akt) (1:1,000; catalog no. 4060) primary
antibodies were obtained from Cell Signaling Technology, Inc.
(Danvers, MA, USA). Rabbit polyclonal anti-EGFR (1:1,000; catalog
no. 18986-1-AP) and rabbit polyclonal anti-β-actin (1:2,000;
catalog no. 20536-1-AP) primary antibodies, and
peroxidase-conjugated AffiniPure goat anti-rabbit/mouse
immunoglobulin G (IgG) secondary antibodies (1:10,000; catalog nos.
SA00001-2 and SA00001-1, respectively) were obtained from
ProteinTech Group, Inc. (Chicago, IL, USA). EasyScript Two-Step
RT-PCR SuperMix kit was obtained from Beijing TransGen Biotech Co.,
Ltd. (Beijing, China). PrimeScript™ RT Reagent kit (catalog no.
RR047A) and SYBR® Premix Ex Taq™ II (catalog no. RR820A)
were obtained from Takara Biotechnology Co., Ltd. (Dalian, China).
Propidium iodide (PI), RNase A, rhodamine 123 (Rh123) and Hoechst
33342 were obtained from Sigma-Aldrich (Merck Millipore, Darmstadt,
Germany). Triton X-100 was obtained from Amresco, LLC (Solon, OH,
USA). TRIzol, adenosine triphosphate (ATP) detection kit (catalog
no. S0026B) and the pH fluorescent probe
2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein
acetoxymethyl ester (BCECF-AM; catalog no. S1006) were obtained
from Biyuntian Biotech Co., Ltd. (Shanghai, China). Mollugin
(positive control) was purchased from National Institutes for Food
and Drug Control (Beijing, China). FFJ-5 (the modified compound of
mollugin) was synthesized by Professor Wenyi Kang (College of
Pharmacy, Henan University, Henan, China).
Cell culture and cell viability
assay
HepG2 (human hepatocellular carcinoma) and A549
(human lung cancer) cells were obtained from the American Type
Culture Collection (Manassas, VA, USA). The cells were maintained
in Dulbecco's modified Eagle medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin in a 37°C incubator with 5% CO2.
Cell viability was detected by MTT assay. HepG2 and A549 cells were
seeded in 96-well plates. After 24 h, the cells were exposed to
FFJ-5 for 48 h, using mollugin as a control. Then, the cells were
incubated in fresh medium containing MTT (0.5 mg/ml; Sigma-Aldrich;
Merck Millipore) at 37°C for 4 h, followed by the addition of 150
µl dimethyl sulfoxide to replace the medium. The absorbance values
at a wavelength of 490 nm were read with a microplate reader (Tecan
Group Ltd., Männedorf, Switzerland).
Western blotting
The cells were harvested in radioimmunoprecipitation
assay buffer [50 mM Tris-HCl (pH 8.0), 150 mM sodium chloride, 1.0%
NP-40, 0.5% sodium deoxycholate and 0.1% SDS] with 10 µg/ml of the
protease inhibitor phenylmethane sulfonyl fluoride, and the cell
lysates were obtained by centrifugation at 12,000 × g for 10
min. The protein concentration in the cells lysates was determined
using a bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology, Haimen, China), and equal amounts of protein (40 µg)
were separated by SDS-PAGE. PKM2, EGFR, pEGFR, Akt, pAkt, PARP and
cleaved PARP were separated using an 8% gel. Caspase-3 and cleaved
caspase-3 were separated using a 12% gel. Gels were subsequently
transferred electrophoretically to a polyvinylidene difluoride
membrane (Merck Millipore) at 70 mA for 2 h. Then, the membrane was
blocked in 5% fat-free milk, and probed with specific primary
antibodies against PKM2, EGFR, pEGFR, Akt, pAkt, caspase-3, cleaved
caspase-3, PARP and cleaved PARP at 4°C overnight. Upon washing in
PBS-Tween 20, the membranes were incubated with anti-mouse IgG or
anti-rabbit IgG secondary antibodies for 2 h at room temperature.
Bands were visualized using an EasyBlot Enhanced Chemiluminescence
kit (Sangon Biotech Co., Ltd., shanghai, China) and detected using
a FluorChem Q Multifluor System (ProteinSimple, San Jose, CA, USA).
β-actin was used as a loading control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated using TRIzol.
Semi-quantitative PCR was performed using an EasyScript Two-Step
RT-PCR SuperMix kit, according to the manufacturer's protocol. A
total of 2 µg RNA was transcribed into complementary DNA with
PrimeScript RT Reagent kit. RT-qPCR was performed with SYBR Premix
Ex Taq II, and the PikoReal™ Real-Time PCR System (Thermo Fisher
Scientific, Inc.) was used to measure messenger RNA (mRNA)
expression. The following thermocycling conditions were used: 95°C
for 5 min; 40 cycles of 95°C for 10 sec, 59°C for 30 sec and 72°C
for 30 sec; 60°C for 30 sec. The reactions for each sample-primer
set were performed in triplicate. Relative quantification analysis
was performed using the comparative Cq (2−ΔΔCq) method
(11). All data were normalized to
the internal control GAPDH. The sequences of the qPCR primers were
as follows: PKM2, forward 5′-AGAACTTGTGCGAGCCTCAA-3′ and reverse
5′-GAGCAGACCTGCCAGACTC-3′ (product length, 128 bp); and GAPDH,
forward 5′-CTCTGCTCCTCCTGTTCGAC-3′ and reverse
5′-ACCAAATCCGTTGACTCCGA-3′ (product length, 109 bp).
ATP production detection
ATP production was detected using the ATP detection
kit, according to the manufacturer's protocol. HepG2 and A549 cells
were seeded in 6-well plates. After 12 h, the cells were treated
with FFJ-5 (0, 10, 20 or 40 µM) for 48 h. Then, the cells were
washed twice with ice-cold PBS and lysed with ATP lysis buffer.
Upon centrifugation at 12,000 × g for 5 min at 4°C to remove
cell debris, the supernatant was added to the substrate solution,
and the product of the reaction (ATP) was measured using a
luminometer (EnSpire®; PerkinElmer, Inc., Waltham, MA,
USA) by calibration with ATP standards and total protein samples.
The ATP production was represented as nm/mg protein.
Intracellular pH detection
Intracellular pH was analyzed using a confocal
scanning laser microscope (FV100-IX81 inverted microscope; Olympus
Corporation, Tokyo, Japan). HepG2 and A549 cells were seeded in
cell culture dishes for confocal microscopy (glass bottom, 20-mm
diameter). After 12 h, the cells were treated with FFJ-5 (0, 10, 20
and 40 µM) for 48 h, and then co-cultured with BCECF-AM (10 µM
diluted in culture medium) for 1 h at 37°C in an incubator with 5%
CO2. The cells were subsequently washed with PBS, and
the PBS was discarded prior to fluorescence determination.
Fluorescence intensity was measured by confocal scanning laser
microscopy at 488 nm excitation and 535 nm emission. The FV10-ASW
software (version 4.2; Olympus Corporation) incorporated in the
confocal microscope was used to calculate the fluorescence
intensity per µm2 automatically, and finally calculated
the relative fluorescence intensity of the samples compared with
that of the control. Changes in relative fluorescence intensity
represent changes in intracellular pH (12).
Cell cycle analysis
Cell cycle analysis was performed using flow
cytometry. HepG2 and A549 cells treated with FFJ-5 (0, 10, 20 or 40
µM) for 24 h were collected and washed twice with ice-cold PBS.
Cells (1×106) were fixed in 75% ethanol at 4°C for ≥4 h.
Cells were then washed twice with ice-cold PBS, followed by
incubation with DNA staining solution [PI (50 µg/ml), RNase A (50
µg/ml) and Triton X-100 (0.5%)] for 20 min. Cell cycle analysis was
immediately performed using flow cytometry (FACSCalibur; BD
Biosciences, Franklin Lakes, NJ, USA).
Cell apoptosis analysis
Cell apoptosis was analyzed using an ArrayScan VTI
HCS 600-type high-content live cell imaging system (Thermo Fisher
Scientific, Inc.). HepG2 and A549 cells were seeded in 96-well
plates. After 12 h, the cells were treated with FFJ-5 (0, 10, 20
and 40 µM) for 48 h, and then washed with ice-cold PBS. Cells were
next incubated with 5 mg/ml Hoechst 33342 for 30 min, and then
incubated with 5 mg/ml PI for a further 1 h at 37°C. The cells were
subsequently washed with ice-cold PBS, and cell apoptosis analysis
was immediately performed using the aforementioned Array Scan VTI
HCS 600-type high-content live cell imaging system. The second
channel, which recorded the overall average fluorescence intensity
(mean fluorescence intensity), represented the number of cells
undergoing late apoptosis.
Mitochondrial membrane potential (MMP)
detection
The change in MMP was analyzed using Rh123 staining,
and the Rh123 content was measured by confocal scanning laser
microscopy (FV100-IX81 inverted microscope; Olympus Corporation).
HepG2 and A549 cells were seeded in cell culture dishes for
confocal microscopy (glass-bottom, 20-mm diameter). After 12 h, the
cells were treated with FFJ-5 (0, 10, 20 and 40 µM) for 48 h, and
then incubated with Rh123 (5 µg/ml) for 30 min at 37°C in and
incubator with 5% CO2. The cells were subsequently
washed with PBS, and the PBS was discarded prior to measurement of
the fluorescence intensity at 488 nm excitation and 535 nm
emission. The aforementioned FV10-ASW software was used to
calculate the fluorescence intensity per µm2
automatically, and finally calculated the relative fluorescence
intensity of the samples compared with that of the control. Changes
in relative fluorescence intensity represent changes in MMP
(13).
Statistical analysis
Each experiment was repeated three times. Relative
cell viability is expressed as a percentage relative to that of the
untreated control cells. Error bars represent standard deviation.
Data were analyzed using analysis of variance for each two-group
comparison test. Statistical analyses were conducted using SPSS
17.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
FFJ-5 inhibits cancer cells
growth
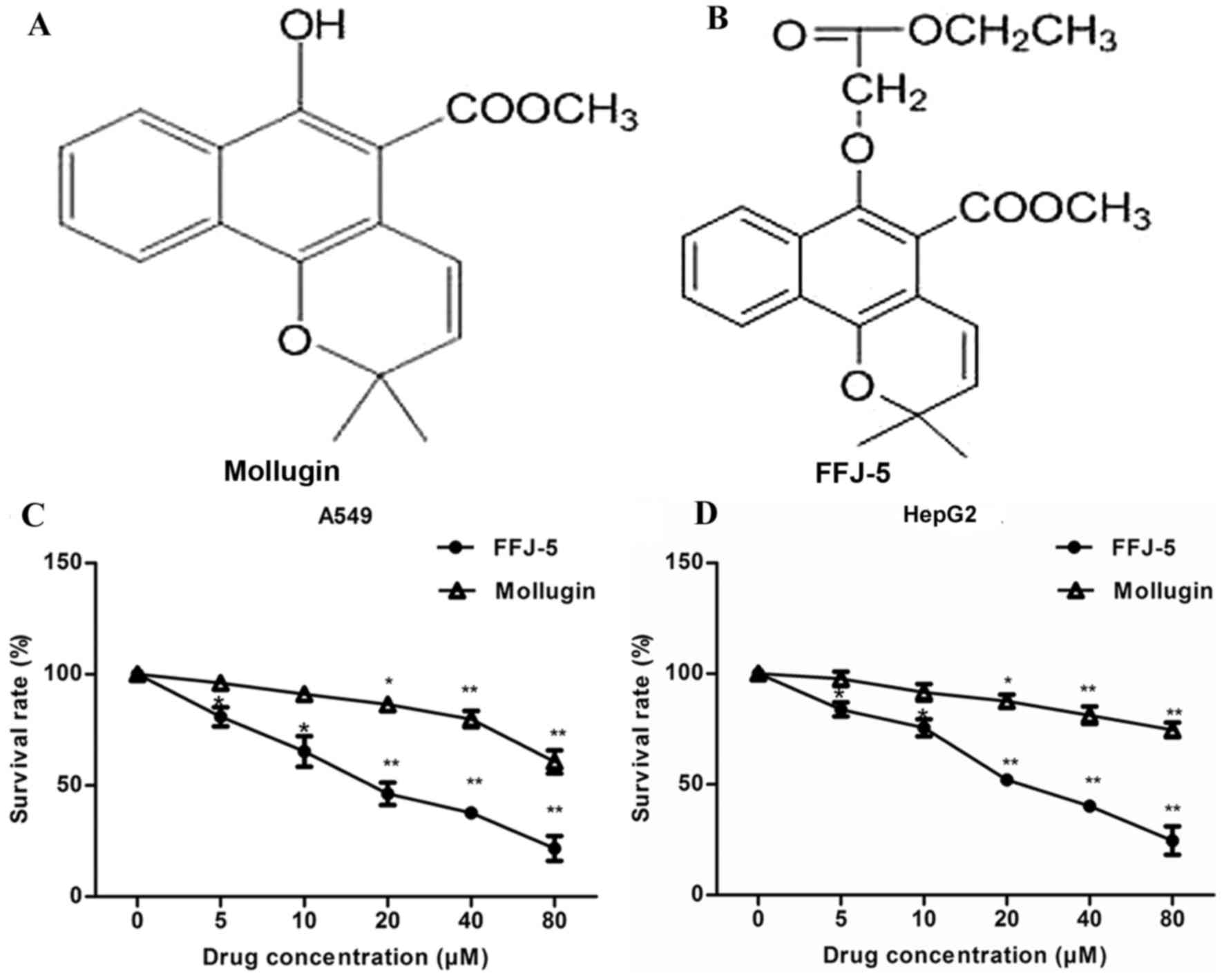
FFJ-5, a novel naphthoquinone derivative, is a
chemically modified compound based on the structure of mollugin
(2) (Fig.
1A and B). It was hypothesized that FFJ-5 possessed stronger
anticancer activity compared with that of mollugin in human cancer
cells. The antiproliferative/survival activity of FFJ-5 in human
liver cancer HepG2 cells and human lung cancer A549 cells was
investigated and compared with that of mollugin. FFJ-5 induced a
significant dose-dependent decrease in cancer cell survival and
exhibited a stronger anticancer activity than that of mollugin in
HepG2 and A549 cells (Fig. 1C and D).
The dose of FFJ-5 required to achieve 50% cell viability was 25.67
and 20.18 µM in HepG2 and A549 cells, respectively.
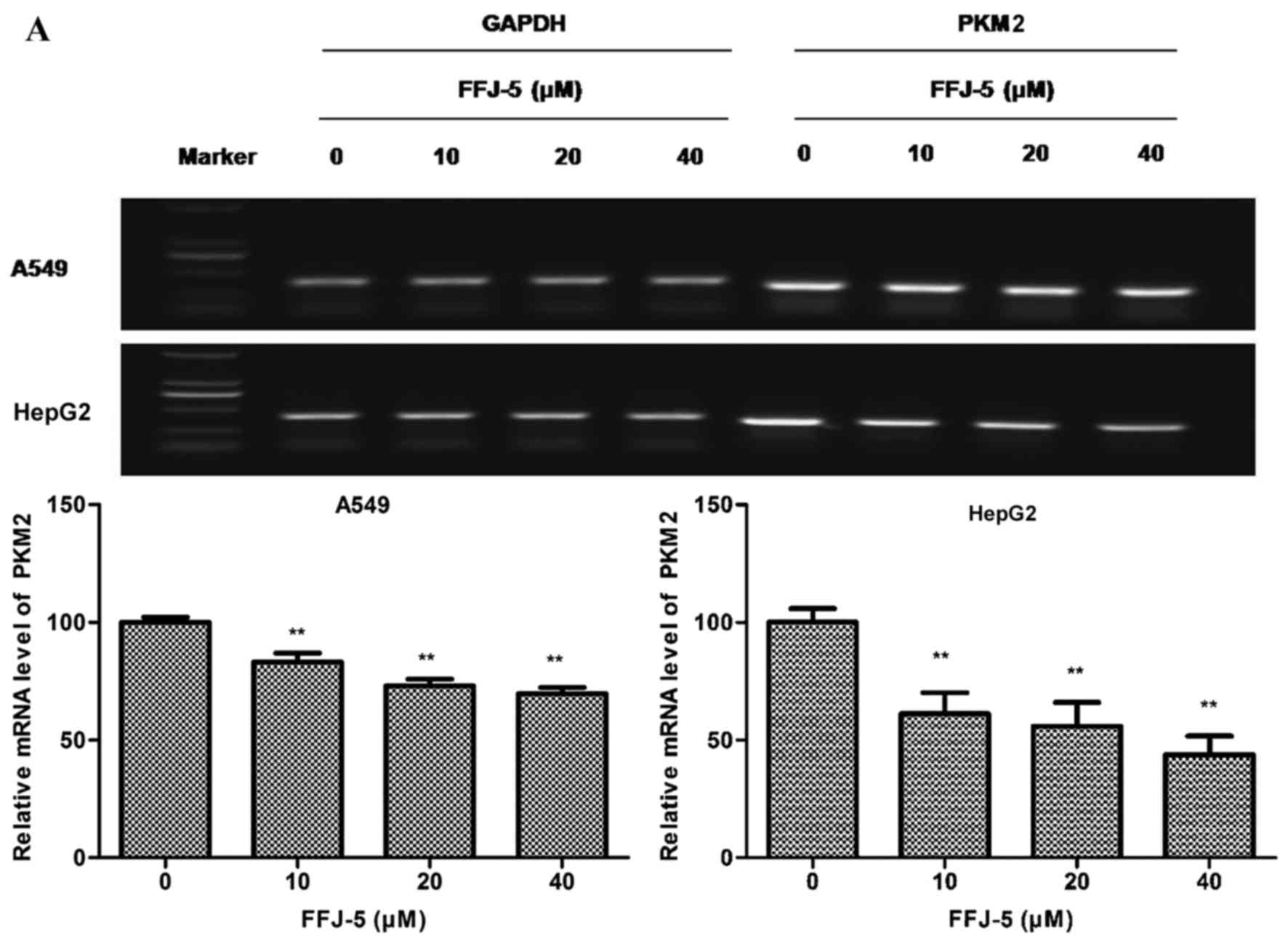
FFJ-5 downregulates the expression of
PKM2
Previous studies had indicated that naphthoquinone
derivatives served as specific inhibitors of PKM2 (14). Therefore, in the present study, the
effect of FFJ-5 on the mRNA and protein levels of PKM2 was measured
in HepG2 and A549 cells. Notably, decreased PKM2 mRNA and protein
levels were observed in HepG2 and A549 cells exposed to FFJ-5, in a
dose-dependent manner (Fig. 2A and
B). In addition, decreased ATP production was observed in HepG2
and A549 cells treated with FFJ-5 (Fig.
2C). These results suggested that FFJ-5 was a valid inhibitor
of PKM2, and consequently inhibited cancer cell growth via
controlling glycolysis and ATP production in cancer cells.
FFJ-5 inhibits the expression and
phosphorylation of EGFR and Akt
Previous studies indicated that the overexpression
of PKM2 in solid tumors was due to the activation of EGFR (15). The authors also demonstrated that EGFR
induced the nuclear translocation of PKM2 and promoted cell
proliferation in liver cancer cells (16). Previous research revealed that reduced
or enhanced Akt phosphorylation was accompanied by the
downregulation or upregulation of PKM2 protein, respectively
(unpublished data). Therefore, in the present study, the effects of
FFJ-5 on the expression and phosphorylation of EGFR and Akt were
evaluated. A decrease in the total EGFR and Akt protein levels, and
a decrease in the phosphorylation of EGFR and Akt, was observed in
HepG2 and A549 cells exposed to FFJ-5 (Fig. 3). The results indicated that FFJ-5 may
inhibit cancer cell growth via the blocking of the EGFR-Akt-PKM2
signaling pathway or via the synergistic action of EGFR, Akt and
PKM2 proteins.
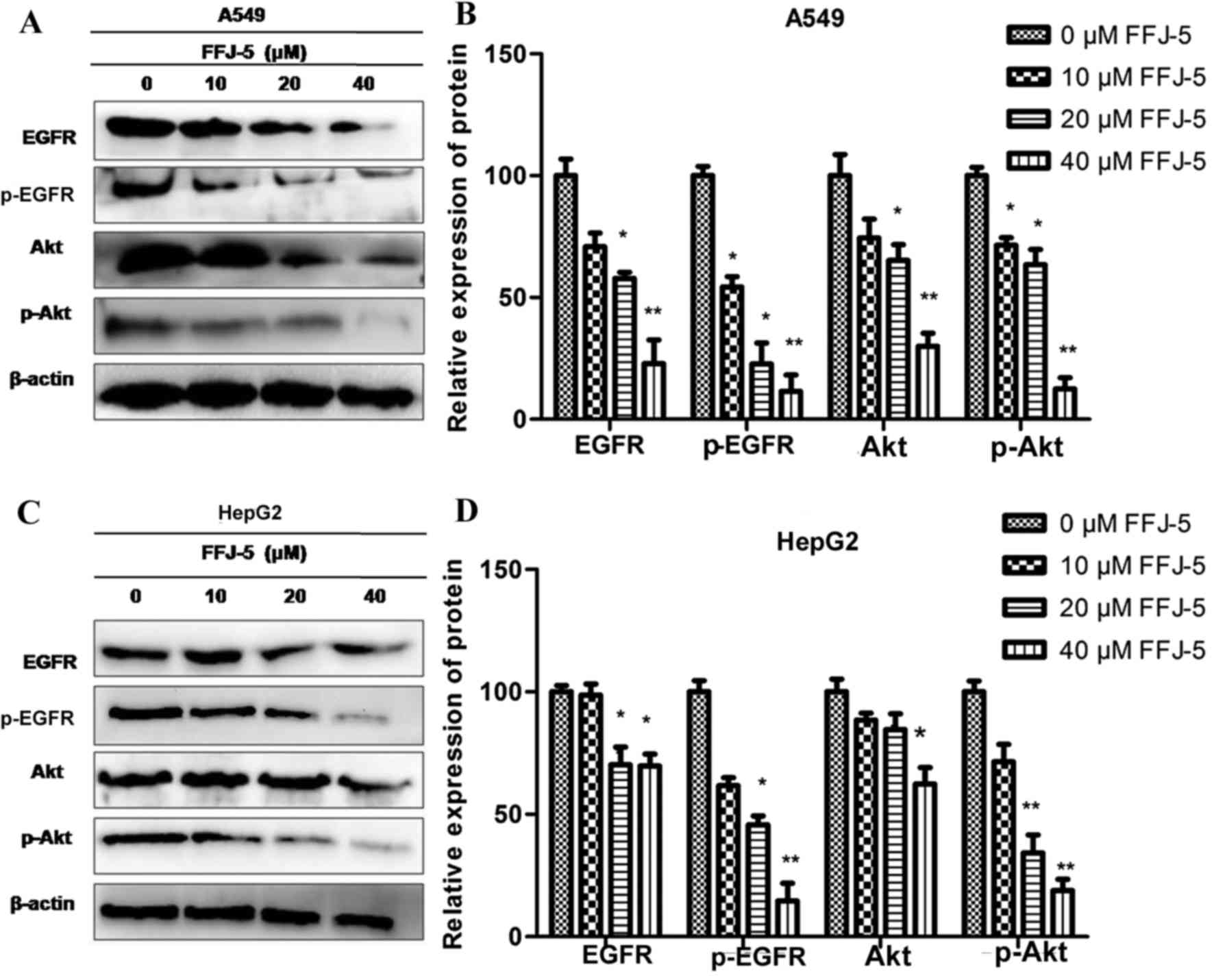 | Figure 3.Effect of FFJ-5 on the expression and
activity of EGFR and Akt. (A) Effect of treatment with different
concentrations of FFJ-5 on the expression of EGFR, pEGFR, Akt and
pAkt in A549 cells. (B) Statistical analysis of the expression
levels of EGFR, pEGFR, Akt and pAkt in A549 cells following
treatment with different concentrations of FFJ-5. (C) Effect of
treatment with different concentrations of FFJ-5 on the expression
of EGFR, pEGFR, Akt and pAkt in HepG2 cells. (D) Statistical
analysis of the expression levels of EGFR, pEGFR, Akt and pAkt in
HepG2 cells following treatment with different concentrations of
FFJ-5. Western blotting revealed that FFJ-5 reduced the total
protein levels of EGFR and Akt, and inhibited the phosphorylation
of and EGFR and Akt. *P<0.05, **P<0.01. EGFR, epidermal
growth factor receptor; p, phosphorylated; Akt, protein kinase
B. |
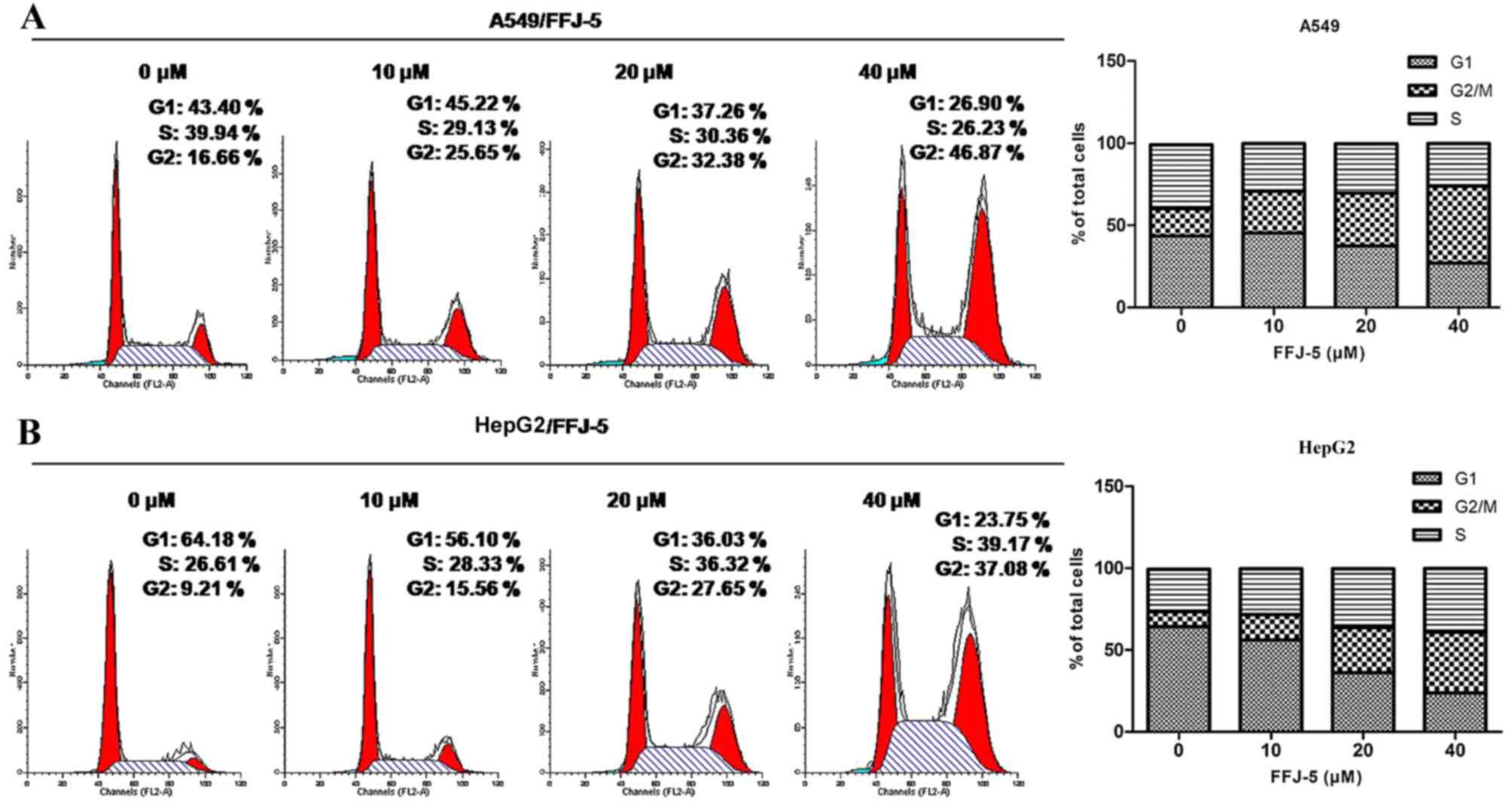
FFJ-5 arrests the cell cycle in the
G2/M phase
Several studies reported that PKM2 in cancer cells
ensured a correct cell division, so that the depletion of PKM2
would cause uneven distribution of the DNA in the two daughter
cells, and consequently would trigger apoptosis in cancer cells
(17). The present study revealed
that FFJ-5 arrested the cell cycle in the G2/M phase in HepG2 and
A549 cells, according to flow cytometry analysis (Fig. 4). These results suggested that FFJ-5
may inhibit cancer cell division due to the inhibition of PKM2 and
consequently cause programmed cell death or apoptosis.
FFJ-5 induces cancer cells
apoptosis
Based on the above results, the possibility that
FFJ-5 caused cancer cell apoptosis was evaluated. Cell apoptosis
was detected with an Array Scan VTI HCS 600-type high-content live
cell imaging system. It was observed that FFJ-5 led to apoptotic
cell death in a dose-dependent manner in HepG2 and A549 cells
(Fig. 5A). In addition, the change in
intracellular pH in cancer cells exposed to FFJ-5 was also studied.
The intracellular pH was verified by confocal scanning laser
microscopy. Decreased intracellular pH level was observed in HepG2
and A549 cells treated with FFJ-5 (Fig.
5B). Additionally, the change in MMP was investigated with
Rh123 staining. Increased Rh123 fluorescence intensity was observed
in HepG2 and A549 cells following FFJ-5 incubation (Fig. 5C). Proteins associated with the
mitochondrial apoptotic pathway were also detected. The western
blot results demonstrated that FFJ-5 activated caspase-3 and
consequently induced the cleavage of PARP (Fig. 5D). Taken together, these results
supported the induction by FFJ-5 of the apoptotic response in HepG2
and A549 cancer cells.
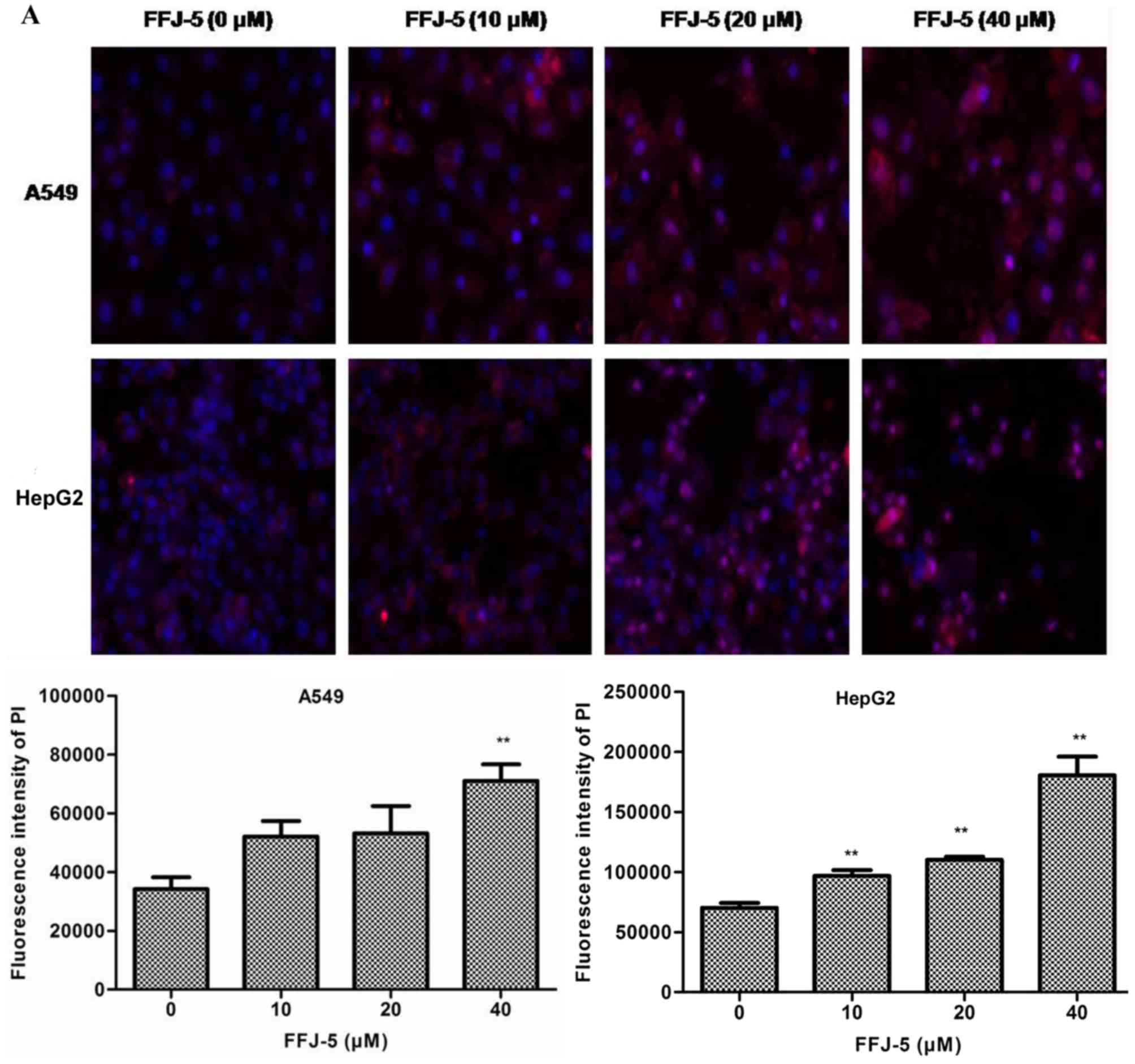 | Figure 5.FFJ-5 reduced the apoptosis of A549
and HepG2 cells. (A) Effect of FFJ-5 on the fluorescence intensity
of PI in A549 and HepG2 cells. Cells treated with 0, 10, 20 and 40
µM FFJ-5 for 48 h were incubated with 5 mg/l Hoechst 33342 for 10
min and with 5 mg/l PI for another 1 h in the dark. Then, the mean
fluorescence intensity of PI was detected with an Array Scan VTI
HCS 600-type high-content live cell imaging system. Magnification,
×20. (B) Effect of FFJ-5 on the intracellular pH level. The
intracellular pH level was verified by confocal scanning laser
microscopy. A decreased intracellular pH level was observed in
HepG2 and A549 cells treated with FFJ-5. The top images are
fluorescent images; the bottom images are light images.
Magnification, ×20. (C) FFJ-5 reduced the MMP in cancer cells. The
change in MMP was detected with Rh123 staining. Increased Rh123
fluorescence intensity in cancer cells upon FFJ-5 incubation was
observed. The top images are fluorescent images; the bottom images
are light images. Magnification, ×20. (D) FFJ-5 activated the
caspase-3 cascade in cancer cells. Caspase-3, cleaved caspase-3,
PARP and cleaved PARP levels were examined in A549 and HepG2 cells
following FFJ-5 treatment for 48 h by western blotting. The data
represent the mean and standard deviation obtained from three
independent experiments. *P<0.05 compared with the
FFJ-5-untreated group. **P<0.01 compared with the
FFJ-5-untreated group. PI, propidium iodide; MMP, mitochondrial
membrane potential; Rh123, rhodamine 123; PARP, poly (ADP-ribose)
polymerase. |
Discussion
Tumor patients mostly present with metabolic
disorders such as high energy consumption and weight loss (18). In the 1920s, Warburg et al
demonstrated that the metabolism of tumor tissues was obviously
enhanced and mainly dependent on glycolytic metabolism (19). Sugar consumption speed in tumor cells
is greater than that in normal cells. Cancer metabolism
characterizes the malignant behavior of tumors. In recent years,
the phenomenon of tumor metabolic abnormalities has gained
attention, and the Warburg effect became the focus of cancer
research (20).
The Warburg effect of cancer cells can promote
cancer cell reproduction and invasion (21). The increase in glycolysis results in
the acidic microenvironment of cancer (22) that is, the extracellular pH of cancer
cells is lower than their intracellular pH, which can enhance the
invasion and proliferation abilities of cancer cells (23). In addition, the acidic
microenvironment of cancer cell metabolism can also sustain cell
proliferation even under conditions of inadequate nutrition
(24). High expression of glycolytic
enzymes can aggravate the acidic microenvironment of cancer cells
(24).
Sugar metabolic enzymes are closely associated with
the cell cycle, and disorders of the cancer cell cycle is one of
the mechanisms of carcinogenesis (25). In 2003, a study indicated that the
metabolic disorder of cancer cells was associated with the cell
cycle regulation of cancer, and that multiple metabolic enzymes
were involved in the regulation of histone H2B genes transcription
during the cell cycle (26).
Therefore, changes in cell metabolism can directly affect the
regulation of the cell cycle.
PKM2, a key metabolic enzyme in cancer cells, can
promote cancer cell's Warburg effect (27). Numerous studies have demonstrated that
PKM2 has a crucial role during the formation and growth of cancer
by increasing the acidic microenvironment and regulating the cell
cycle and cell apoptosis (28,29).
Naphthoquinone derivatives are specific inhibitors of PKM2
(14). FFJ-5, a novel naphthoquinone
derivative, is a chemically modified compound based on the
structure of mollugin (2). In the
present study, high expression of PKM2 was detected in A549 and
HepG2 cells by western blotting, indicating the important role of
glycolysis in the development of lung and liver cancer. Our study
revealed that FFJ-5 inhibited the expression of PKM2 protein,
arrested the cell cycle in the G2/M phase and reduced the ATP
production in A549 and HepG2 cells. Decreased intracellular pH
levels and activated caspase cascade were also observed in cancer
cells exposed to FFJ-5. FFJ-5 inhibited cancer cell growth and
induced cancer cell apoptosis by blocking the EGFR-Akt-PKM2
signaling pathway, or via the synergistic action of EGFR, Akt and
PKM2 proteins. These data suggest that FFJ-5 induced apoptosis in
A549 and HepG2 cells by inhibiting PKM2 expression.
In conclusion, the novel naphthoquinone compound
FFJ-5 induces apoptosis in human lung adenocarcinoma A549 and human
hepatoma HepG2 cells, and may be an effective anti-cancer candidate
agent.
Acknowledgements
The authors would like to acknowledge the financial
assistance provided by a grant from the Key Science and Technology
Fund of Henan Province in China (grant no. 142300410128) to support
the present project.
References
|
1
|
Shilpa PN, Venkatabalasubramanian S and
Devaraj SN: Ameliorative effect of methanol extract of Rubia
cordifolia in N-nitrosodiethylamine-induced hepatocellular
carcinoma. Pharm Biol. 50:376–383. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang L, Wang H, Zhu J, Xu J and Ding K:
Mollugin induces tumor cell apoptosis and autophagy via the
PI3K/AKT/mTOR/p70S6K and ERK signaling pathways. Biochem Biophys
Res Commun. 450:247–254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.PubMed/NCBI
|
|
4
|
Cairns RA: Drivers of the Warburg
phenotype. Cancer J. 21:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koppenol WH, Bounds PL and Dang CV: Otto
Warburg's contributions to current concepts of cancer metabolism.
Nat Rev Cancer. 11:325–337. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu H, Li Z, Wang Y, Yang P, Li Z, Li H and
Wu C: MiR-106b-mediated Mfn2 suppression is critical for PKM2
induced mitochondrial fusion. Am J Cancer Res. 6:2221–2234.
2016.PubMed/NCBI
|
|
8
|
Warner SL, Carpenter KJ and Bearss DJ:
Activators of PKM2 in cancer metabolism. Future Med Chem.
6:1167–1178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iqbal MA, Gupta V, Gopinath P, Mazurek S
and Bamezai RN: Pyruvate kinase M2 and cancer: An updated
assessment. FEBS Lett. 588:2685–2692. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chu B, Wang J, Wang Y and Yang G:
Knockdown of PKM2 induces apoptosis and autophagy in human A549
alveolar adenocarcinoma cells. Mol Med Rep. 12:4358–4363.
2015.PubMed/NCBI
|
|
11
|
Fendri A, Kontos CK, Khabir A,
MokdadGargouri R, Ardavanis A and Scorilas A: Quantitative analysis
of BCL2 mRNA expression in nasopharyngeal carcinoma: An unfavorable
and independent prognostic factor. Tumour Biol. 31:391–399. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen M, Zou X, Luo H, Cao J, Zhang X,
Zhang B and Liu W: Effects and mechanisms of proton pump inhibitors
as a novel chemosensitizer on human gastric adenocarcinoma
(SGC7901) cells. Cell Biol Int. 33:1008–1019. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren J, Cheng H, Xin WQ, Chen X and Hu K:
Induction of apoptosis by 7-piperazinethylchrysin in HCT-116 human
colon cancer cells. Oncol Rep. 28:1719–1726. 2012.PubMed/NCBI
|
|
14
|
Chen J, Jiang Z, Wang B, Wang Y and Hu X:
Vitamin K(3) and K(5) are inhibitors of tumor pyruvate kinase M2.
Cancer Lett. 316:204–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang
L, You MJ, Koh MY, Cote G, Aldape K, et al: EGFR-induced and PKCε
monoubiquitylation-dependent NF-κB activation upregulates PKM2
expression and promotes tumorigenesis. Mol Cell. 48:771–784. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan FT, Shen CS, Tao L, Tian C, Liu ZG,
Zhu ZJ, Liu YP, Pei CS, Wu HY, Zhang L, et al: PKM2 regulates
hepatocellular carcinoma cell epithelial-mesenchymal transition and
migration upon EGFR activation. Asian Pac J Cancer Prev.
15:1961–1970. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang Y, Li X, Yang W, Hawke DH, Zheng Y,
Xia Y, Aldape K, Wei C, Guo F, Chen Y and Lu Z: PKM2 regulates
chromosome segregation and mitosis progression of tumor cells. Mol
Cell. 53:75–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Flint TR, Janowitz T, Connell CM, Roberts
EW, Denton AE, Coll AP, Jodrell DI and Fearon DT: Tumor-induced
IL-6 reprograms host metabolism to suppress anti-tumor immunity.
Cell Metab. 24:672–684. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao JL and Chen YG: Natural compounds
regulate glycolysis in hypoxic tumor microenvironment. Biomed Res
Int. 2015:3541432015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Zhao Z, Zhou Z and Liu R: PKM2
promotes cell survival and invasion under metabolic stress by
enhancing warburg effect in pancreatic ductal adenocarcinoma. Dig
Dis Sci. 61:767–773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Justus CR, Sanderlin EJ and Yang LV:
Molecular connections between cancer cell metabolism and the tumor
microenvironment. Int J Mol Sci. 16:11055–11086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fan SH, Wang YY, Wu ZY, Zhang ZF, Lu J, Li
MQ, Shan Q, Wu DM, Sun CH, Hu B and Zheng YL: AGPAT9 suppresses
cell growth, invasion and metastasis by counteracting acidic tumor
microenvironmentthrough KLF4/LASS2/V-ATPase signaling pathway in
breast cancer. Oncotarget. 6:18406–18417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng J: Energy metabolism of cancer:
Glycolysis versus oxidative phosphorylation (Review). Oncol Lett.
4:1151–1157. 2012.PubMed/NCBI
|
|
25
|
Tan X, Ye H, Yang K, Chen D and Tang H:
Circadian rhythm variation of the clock genes Per1 and cell cycle
related genes in different stages of carcinogenesis of buccal
mucosa in animal model. Zhonghua Kou Qiang Yi Xue Za Zhi.
50:392–398. 2015.(In Chinese). PubMed/NCBI
|
|
26
|
Zheng L, Roeder RG and Luo Y: S phase
activation of the histone H2B promoter by OCA-S, a coactivator
complex that contains GAPDH as a key component. Cell. 114:255–266.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee KM, Nam K, Oh S, Lim J, Lee T and Shin
I: ECM1 promotes the Warburg effect through EGF-mediated activation
of PKM2. Cell Signal. 27:228–235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Papadaki C, Sfakianaki M, Lagoudaki E,
Giagkas G, Ioannidis G, Trypaki M, Tsakalaki E, Voutsina A,
Koutsopoulos A, Mavroudis D, et al: PKM2 as a biomarker for
chemosensitivity to front-line platinum-based chemotherapy in
patients with metastatic non-small-cell lung cancer. Br J Cancer.
111:1757–1764. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen Z, Wang Z, Guo W, Zhang Z, Zhao F,
Zhao Y, Jia D, Ding J, Wang H, Yao M, et al: TRIM35 interacts with
pyruvate kinase isoform M2 to suppress the Warburg effect and
tumorigenicity in hepatocellular carcinoma. Oncogene. 34:3946–3956.
2015. View Article : Google Scholar : PubMed/NCBI
|