Introduction
Breast cancer is the most common type of cancer
among women worldwide, accounting for 29% of novel cancer cases in
2014 (1). A total of 39,620 breast
cancer mortalities were reported among women in 2013 in the USA
despite constant breast cancer incidence (2). Considering the heterogeneity of breast
cancer, diverse terms have been used to explain the underlying
biological and pathological characteristics, responses to therapy
and clinical outcomes (3). The
molecular mechanisms of carcinogenesis are complex due to aberrant
protein expression, gene changes and miRNA deregulation. Therefore,
numerous studies have focused on screening for novel diagnostic and
prognostic biomarkers and therapeutic targets in breast cancer
(4–6).
Polycomb group (PcG) complexes mediate the inherent
stability of cells. These proteins regulate the expression of
numerous genes that control the maintenance, differentiation and
proliferation of adult stem cells and cancer cells (7). Biochemical characterization has
categorized PcG complexes into two subtypes: Polycomb repressive
complex (PRC) 1 and PRC2 (8–10). The chromobox (Cbx) family comprises
five members (Cbx2, Cbx4, Cbx6, Cbx7 and Cbx8) in mammals (11), and it is a component of PRC1. Numerous
studies have indicated that the Cbx family is associated with
cancer. High Cbx7 expression has been found to associate with
ovarian clear cell adenocarcinoma, lymphomagenesis and gastric
cancer (12–14). Cbx4 exerts a critical function in
tumor angiogenesis by controlling the hypoxia-inducible factor-1α
protein (15). However, the
association between Cbx2 expression and cancer remains unclear.
Recent evidence has confirmed that the overexpression of Cbx2
results in the differentiation and exhaustion of hematopoietic stem
cells (16). Notably, a number of
malignant tumors with normal gene copy numbers demonstrate
recurrent Cbx2 overexpression (17).
In the present study, Cbx2 protein expression was
analyzed by immunohistochemistry (IHC) using tissue microarrays
(TMAs) in an independent cohort of patients with breast cancer.
Furthermore, the association between Cbx2 expression and the
clinicopathological features, survival and chemotherapy outcomes of
breast cancer patients were analyzed. The aim of the study was to
determine whether Cbx2 presents a potential prognostic marker and
alternative therapeutic target in breast cancer.
Materials and methods
Patients and clinical samples
A total of 455 patients primarily diagnosed with
breast cancer, who underwent surgery at The Affiliated Tumor
Hospital of Harbin Medical University (Harbin, China) between March
and December in 2006, were consecutively recruited for the present
study. None of the patients had received any treatment prior to
surgery. All patients were pathologically diagnosed with invasive
ductal cancer according to the World Health Organization
classification of breast tumors (18)
and the median age of patients was 49 years (range, 25–78 years).
Patients with a previous history of tumors, including recurrent
tumors, metastatic disease and bilateral tumors, and patients who
had previously received neoadjuvant treatment were excluded. A
total of 455 tumor tissue specimens and 216 corresponding adjacent
normal tissues located 5 cm from the cancer margin were archived
from the Department of Pathology at the Affiliated Tumor Hospital
of Harbin Medical University. A total of 216 paired tumor and
normal breast tissues and 239 unpaired tumor tissues were collected
for further study. Patient information regarding tumor size,
pathological grade, lymph node status and chemotherapy treatment
were obtained from medical records. Among the 309 patients with
complete records of adjuvant chemotherapy regimen, 7 accepted Taxol
cis-platinum treatment, 13 accepted taxol fluorouracil treatment,
43 accepted Taxol epirubicin treatment, 9 accepted nedaplatin
cis-platinum treatment, 10 accepted nedaplatin fluorouracil
treatment, 22 accepted nedaplatin epirubicin treatment, 143
accepted fluorouracil epirubicin cyclophosphamide treatment, 28
accepted epirubicin Taxol treatment and 34 accepted more than two
regimens. According to regimens including Taxol, patients were
split into two groups. One group contained 122 patients who
received chemotherapy, including Taxol, and another group contained
187 patients who received chemotherapy without Taxol. All patients
provided written informed consent for the use of their clinical
specimens for medical research. This study was approved by the
research medical ethics committee of Harbin Medical University.
Histology, TMA and IHC
The tissues obtained from surgical removal were
rapidly fixed in 10% neutral buffered formalin. Subsequent to
dehydration, clearing, infiltration and paraffin-embedding, the
prepared tissue blocks were cut into 4-mm sections for hematoxylin
and eosin staining. Three cores (2 mm in diameter) were obtained
from each breast cancer sample and inserted into the recipient TMA
blocks. A total of 216 invasive ductal carcinomas and corresponding
normal breast tissue samples were inserted into three TMA blocks,
and 239 unpaired cancer tissues were fixed in two TMA blocks. All
TMA blocks were cut with a microtome to 4-µm sections and affixed
to a slide treated with 5% poly-lysine.
Estrogen receptor (ER), progesterone receptor (PR),
human epidermal growth factor receptor-2 (HER-2) and tumor protein
53 (p53) expression and Ki-67 were routinely assayed by IHC, as
previously described (19). Briefly,
IHC staining was performed for ER and PR using 4-µm paraffin
sections cut from TMA blocks. ER, PR and HER-2 markers were
immunostained in a single process using hematoxylin and eosin
stains and the following primary monoclonal antibodies: Mouse
anti-human ER (1:100; ZM-0104), mouse anti-human PR (1:150;
ZM-0215), mouse anti-human HER2 (1:100; ZM-0065) (Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd., Beijing, China), mouse
anti-human Ki-67 (1:100; IR62661; Dako, Glostrup, Denmark) and
mouse anti-human p53 (1:400; sc-47698; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) antibodies. Samples were incubated with the
primary antibodies overnight at 4°C, followed by incubation with a
biotin-labeled goat anti-mouse immunoglobulin (Ig) G secondary
antibody (SP-9002; Beijing Zhongshan Golden Bridge Biotechnology
Co., Ltd.) at room temperature for 30 min. Fluorescence in
situ hybridization (FISH) assays were performed to determine
HER-2 status in tumors with 2+ immunoreactivity according to
guidelines (20). Tumor cells were
considered to exhibit positive ER and PR expression when >10% of
the tumor cell nuclei were stained in the three cores. Tumor cells
were considered to exhibit HER-2 protein overexpression when
>10% of cells exhibited strong membrane staining (3+) or
positive signals in the FISH tests (21). The Ki-67 score was defined as the
percentage of positively stained cells, regardless of the
intensity, among the total number of invasive cells in the scored
area (22). Positive staining for
Ki-67 was defined when >10% of stained cells exhibited
positivity. For p53, positive staining of <10% of tumor cells
was defined as negative tumor expression, whereas staining of ≥10%
tumor cells indicated positive tumor expression (23,24).
Cbx2 protein expression was evaluated using
immunostained TMA slides from each core. IHC was conducted as
follows: Briefly, antigen retrieval was performed using 10 mM
sodium citrate (pH 6.0) and sections were washed with Tris-buffered
saline. To block endogenous peroxidase activity, the sections were
treated with 3% H2O2 for 10 min. Non-specific
binding was blocked by incubation with 1% low lethal serum (Boster
Inc., Wuhan, China) in PBS. Next, the slides were incubated with
anti-CBX2 polyclonal antibodies (1:300; PA5-309961; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 4°C overnight. The slides
were then incubated with goat anti-rabbit IgG secondary antibody
(SP-9001; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.)
at room temperature for 30 min. After washing with PBS three times,
each section was treated with 300–500 ml diaminobenzidine working
solution at room temperature for 5–10 min for visualization and
then washed with distilled water.
IHC evaluation of Cbx2 protein
expression
Immunostaining was evaluated by two breast
pathologists from the The Affiliated Tumor Hospital of Harbin
Medical University, who were blinded to the patient clinical
outcomes. Scoring was performed using the semi-quantitative score
method (25) to calculate the product
of the percentage and intensity of positively stained tumor cells
within the invasive tissue component. In the cytoplasm, staining
intensity was graded as follows: 0, no staining; 1, weak staining
(light yellow); 2, moderate staining (yellow brown); and 3, strong
staining (brown). The percentage (0–100%) of staining was scored as
follows: 0, no positive tumor cells; 1, <25% positive tumor
cells; 2, 25–50% positive tumor cells; 3, 51–75% positive tumor
cells; and 5, >75% positive tumor cells (25,26). The
immunoreactive score (IRS) ranged between 0 and 12. An IRS score
(IRS = staining percentage × staining intensity) of <6 was
classified as low expression, whereas a score of >6 indicated
high expression.
Follow-up
All patients were advised to attend follow-up
examinations every 4–6 months for the first 5 years following
surgery, and every 12 months thereafter. All patients were
regularly followed up until mortality or the study end date (30
December, 2012). Prognosis was recorded by the Center of Medical
Records at The Affiliated Tumor Hospital of Harbin Medical
University. Overall survival (OS) time was assessed for prognostic
analysis.
Statistical and survival analyses
All statistical analyses were performed using SPSS
13.0 statistical software (SPSS, Inc., Chicago, IL, USA). The
difference in Cbx2 expression between breast cancer tissues and
normal tissues was assessed using the Mann-Whitney U test. The
association between Cbx2 and patient clinicopathological features
was analyzed using the χ2 test. OS time was determined
as the time from surgery to the date of mortality or last
follow-up. Kaplan-Meier analysis was used to estimate OS time.
Univariate analysis was performed using the log-rank test.
Univariate and multivariate Cox proportional hazard models were
used to assess clinicopathological prognostic factors affecting OS.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Cbx2 protein expression in cancer and
normal breast tissues
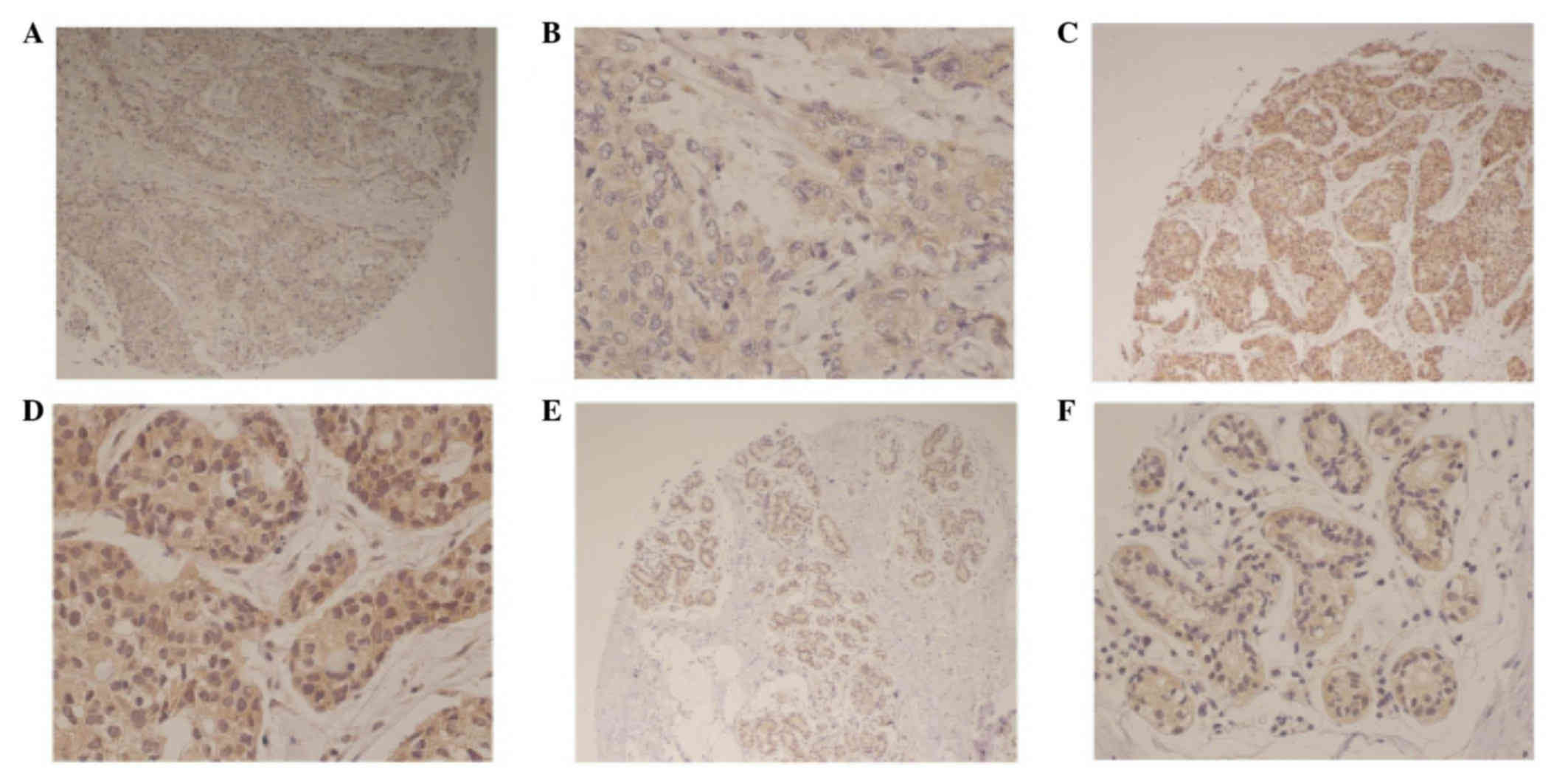
Cbx2 expression was identified in the cytoplasm of
breast cancer tissues (Fig. 1). Cbx2
expression was also identified in the cytoplasm of matched adjacent
normal tissues (Fig. 1A and B).
Representative immunohistochemical images demonstrate low and high
Cbx2 expression (Fig. 1C-F).
A total of 216 paired cancer tissues and matched
adjacent normal tissues were obtained for analysis. Cbx2 protein
expression was identified in 199/216 tumor tissues and 196/216
adjacent tissues. Notably, in 15 (6.94%) paired cancer and adjacent
normal tissues, Cbx2 expression was higher in normal tissues
compared with cancer tissues. However, in 175 (81.01%) normal
tissues, Cbx2 expression was lower compared with in cancer tissues.
A total of 26 (12.04%) paired tissues exhibited equal Cbx2
expression. The median and mean IRS of 455 tumor tissues were 6.00
and 6.21, respectively, whereas the median and mean IRS of the 216
matched normal adjacent tissues were 3.00 and 3.48, respectively.
The protein expression of Cbx2 was significantly higher in tumor
tissues compared with adjacent normal tissues (P<0.001). Using
an IRS of 6 as the cut-off value for high Cxb2 expression, 53.89%
(245/416) tumor tissues and 11.11% (24/216) adjacent normal tissues
exhibited high Cbx2 expression (P<0.001).
Association between Cbx2 protein
expression and patient clinicopathological features
A total of 455 tumor tissues were included in the
present study. The median IRS of Cbx2 expression was 6, which was
used as a cut-off for high expression. Based on this cut-off value,
46.15% (210/455) and 53.85% (245/455) tumor tissues exhibited low
and high Cbx2 cytoplasmic expression, respectively. The expression
of cytoplasmic Cbx2 was found to significantly associate with tumor
size (P<0.001), lymph node metastasis (P=0.008), tumor node
metastasis (TNM) classification of malignant tumors (18) stage (P<0.001) and positive HER-2
status (P=0.048) (Table I).
 | Table I.Association between Cbx2 expression
and patient clinicopathological features. |
Table I.
Association between Cbx2 expression
and patient clinicopathological features.
|
|
| Cbx2
expression |
|
|---|
|
|
|
|
|
|---|
| Parameter | n | Low, n (%) | High, n (%) | P-value |
|---|
| Age, years |
|
|
| 0.441 |
|
<50 | 249 | 119 (47.8) | 130 (52.2) |
|
|
≥50 | 206 | 91
(44.2) | 115 (55.8) |
|
| Tumor size, cm |
|
|
| <0.001 |
|
<2 | 101 | 65
(64.4) | 36
(35.6) |
|
| ≥2 | 353 | 144 (40.8) | 209 (59.2) |
|
| Pathological
stage |
|
|
| 0.286 |
| I | 40 | 23
(57.5) | 17
(42.5) |
|
| II | 121 | 56
(46.3) | 65
(53.7) |
|
|
III | 285 | 126 (44.2) | 159 (55.8) |
|
| LNM |
|
|
| 0.008 |
|
Negative | 210 | 111 (52.9) | 99
(47.1) |
|
|
Positive | 245 | 99
(40.4) | 146 (59.6) |
|
| TNM stage |
|
|
| <0.001 |
| I | 91 | 75
(82.4) | 16
(17.6) |
|
| II | 225 | 131 (58.2) | 94
(41.8) |
|
|
III | 139 | 4
(2.9) | 135 (97.1) |
|
| Ki-67, % |
|
|
| 0.080 |
|
<10 | 147 | 77
(52.4) | 70
(47.6) |
|
|
≥10 | 305 | 133 (43.6) | 172 (56.4) |
|
| HER-2 status |
|
|
| 0.048 |
|
Negative | 363 | 176 (48.5) | 187 (51.5) |
|
|
Positive | 92 | 34
(37.0) | 58
(63.0) |
|
| ER status |
|
|
| 0.162 |
|
Negative | 242 | 104 (43.0) | 138 (57.0) |
|
|
Positive | 212 | 105 (49.5) | 107 (50.5) |
|
| PR status |
|
|
| 0.174 |
|
Negative | 180 | 76
(42.2) | 104 (57.8) |
|
|
Positive | 275 | 134 (48.7) | 141 (51.3) |
|
| p53 status |
|
|
| 0.717 |
|
Negative | 79 | 35
(44.3) | 44
(55.7) |
|
|
Positive | 376 | 175 (46.5) | 201 (53.5) |
|
| Subtype |
|
|
| 0.388 |
|
HER-2 | 92 |
37(40.2) | 55
(59.8) |
|
| Luminal
A | 85 | 44
(51.8) | 41
(48.2) |
|
| Luminal
B | 234 | 111 (47.4) | 123 (52.6) |
|
| Triple
negative | 44 | 18
(40.9) | 26
(59.1) |
|
Association between Cbx2 expression
and prognosis of patients with breast cancer
A total of 403 patients were followed up and 52
patients were lost to follow-up. During the follow-up period,
10.93% (20/183) and 18.64% (41/220) patients in the low and high
Cbx2 expression groups succumbed to the disease, respectively. The
mean survival times were 77.37 months (range, 75.66–79.07 months)
and 74.29 months (range, 71.94–76.64 months) in the low and high
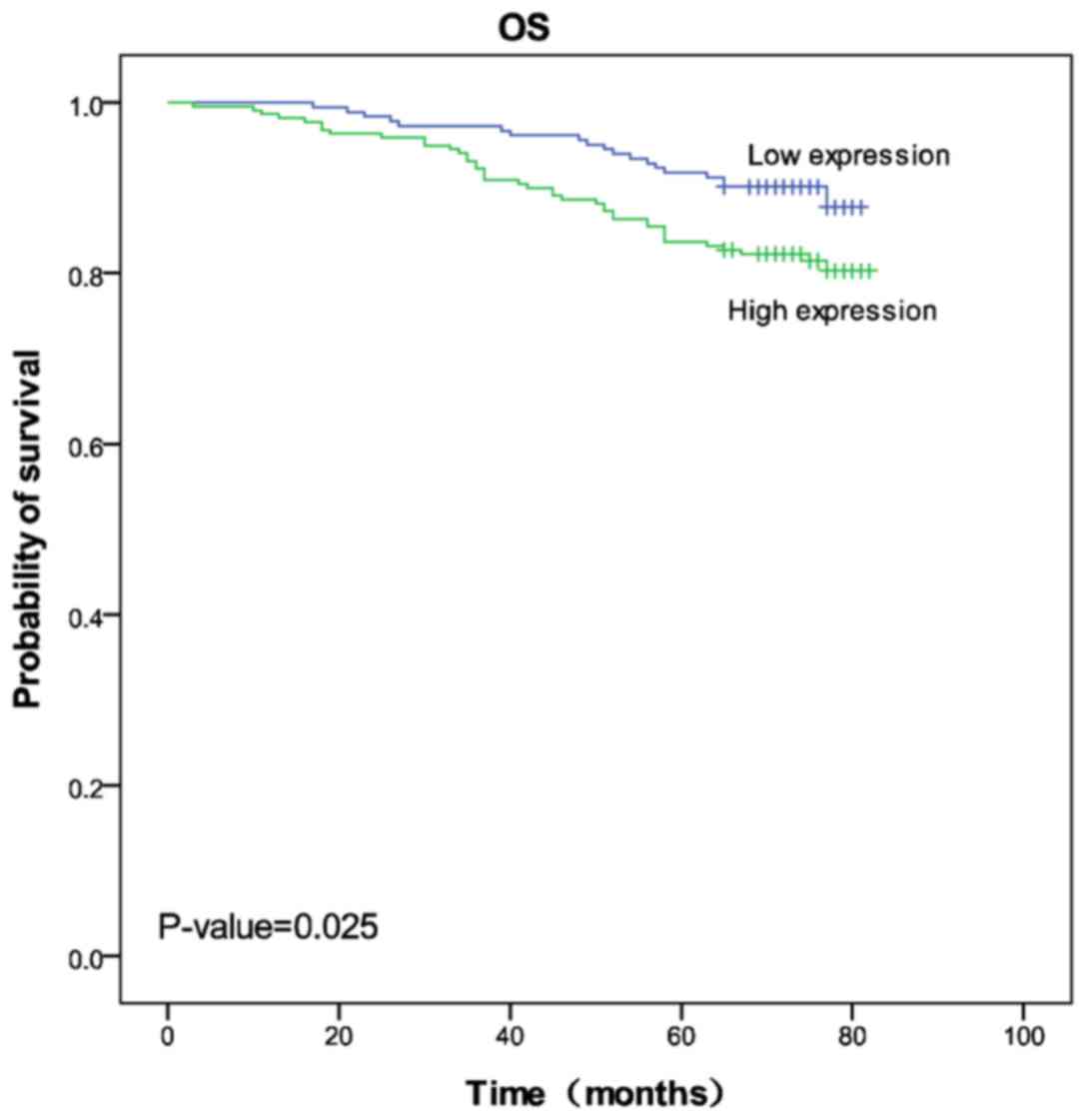
Cbx2 expression groups, respectively. Kaplan-Meier 5-year survival
curves were stratified for Cbx2 expression and the results revealed
that high Cbx2 expression was associated with poor prognosis
(log-rank test statistic, 5.032; P=0.025; Fig. 2).
Univariate and multivariate Cox regression analyses
were performed to evaluate the association between Cbx2 expression
and clinicopathological features on patient prognosis. Variables
such as tumor size, pathological stage, lymph node metastasis, TNM
stage, Ki-67 status, PR status and molecular subtype were included
in multivariate analyses, which were associated with survival of
patients with breast cancer as identified by the log-rank test.
Univariate Cox analysis demonstrated significantly shorter OS time
in patients with a large tumor size [(hazard ratio (HR), 2.361; 95%
confidence interval (CI), 1.074–5.189; P=0.033], positive PR status
(HR, 0.579; 95% CI, 0.350–0.956; P=0.033), positive Ki-67 status
(HR, 1.920; 95% CI, 1.040–3.545; P=0.037) and high Cbx2 expression
(HR, 1.826; 95% CI, 1.069–3.116; P=0.027) (Table II). The multivariate Cox proportional
hazard model revealed that only high TNM stage (HR=3.427; 95%
CI=1.363–8.614; P=0.009) was independently associated with poor
survival (Table II).
 | Table II.Univariate and multivariate cox
regression analyses of overall survival time in breast cancer
patients. |
Table II.
Univariate and multivariate cox
regression analyses of overall survival time in breast cancer
patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Tumor size, cm
(≥2/<2) | 2.361 | 1.074–5.189 | 0.033 | 1.231 | 0.542–2.795 | 0.619 |
| Pathological stage
(I/II/III) | 1.778 | 1.095–2.888 | 0.020 | 1.399 | 0.838–2.336 | 0.199 |
| TNM stage
(positive/negative) | 3.671 | 1.989–6.777 | <0.001 | 3.427 | 1.363–8.614 | 0.009 |
| LNM
(positive/negative) | 3.069 | 2.006–4.696 | <0.001 | 1.025 | 0.389–2.699 | 0.960 |
| Ki-67 status
(positive/negative) | 1.920 | 1.040–3.545 | 0.037 | 1.673 | 0.866–3.233 | 0.126 |
| PR status
(positive/negative) | 0.579 | 0.350–0.956 | 0.033 | 0.878 | 0.333–2.314 | 0.792 |
| Subtype |
|
|
|
|
|
|
| HER-2
status (positive/negative) | Reference |
|
| Reference |
|
|
| Luminal
A | 0.878 | 0.433–1.781 | 0.718 | 1.264 | 0.409–3.909 | 0.684 |
| Luminal
B | 0.488 | 0.259–0.919 | 0.026 | 0.698 | 0.250–1.948 | 0.493 |
| Triple
negative | 1.108 | 0.478–2.569 | 0.810 | 1.608 | 0.677–3.823 | 0.282 |
| Cbx2 expression
(high/low) | 1.826 | 1.069–3.116 | 0.027 | 1.790 | 1.048–3.056 | 0.236 |
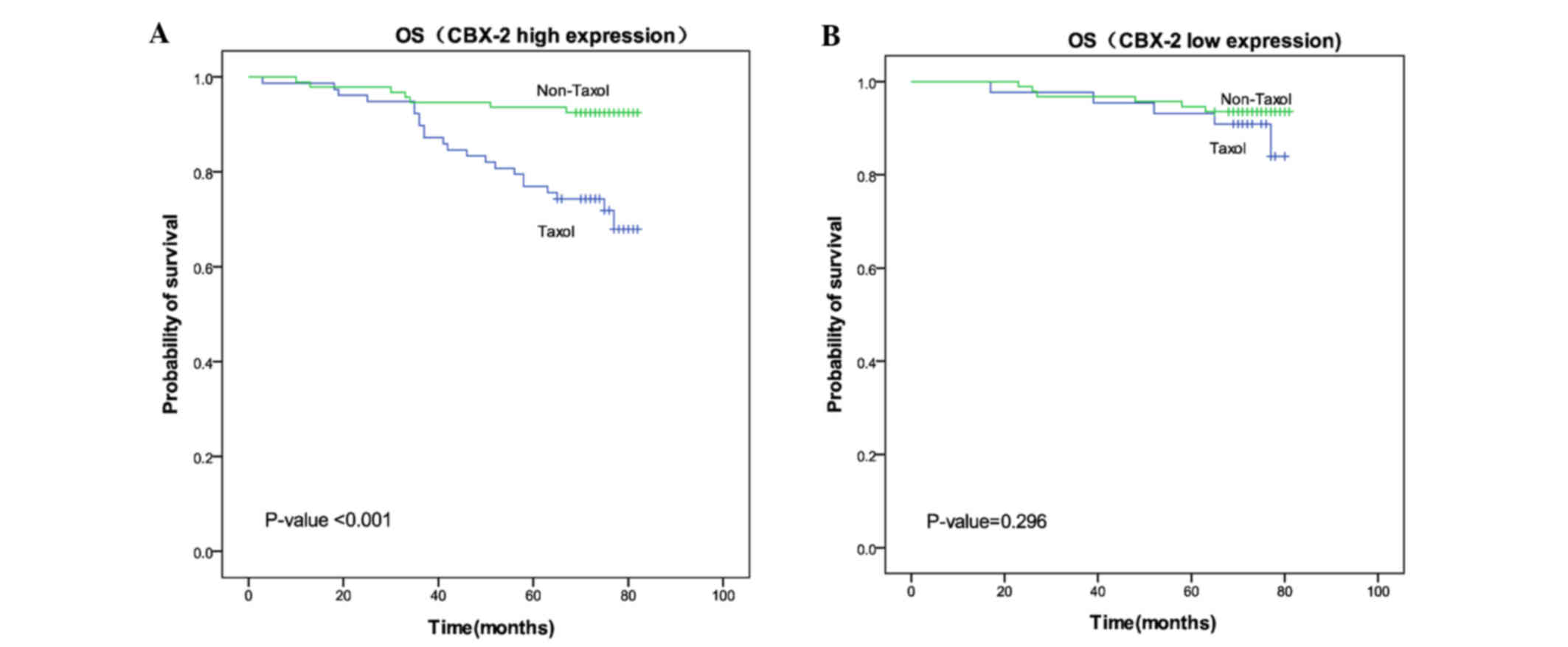
Effect of Taxol in patients expressing
Cbx2
A total of 309 patients underwent chemotherapy
treatment. Among these patients, 122 received Taxol and 187
received an alternative chemotherapy regimen without Taxol
(including nedaplatin cis-platinum, nedaplatin fluorouracil,
nedaplatin epirubicin and fluorouracil epirubicin
cyclophosphamide). Among patients with high Cbx2 expression, the
mean OS time of patients receiving Taxol treatment (71.01 months)
was significantly shorter compared with patients receiving
treatment without Taxol (78.43 months) (P<0.001). However, in
the low Cbx2 expression group, no significant difference in the
mean OS time of patients was identified between those treated with
Taxol (76.45 months) and those without Taxol treatment (78.41
months) (P=0.296). These results indicated that patients with high
Cbx2 expression do not exhibit sensitivity to chemotherapy programs
that include Taxol (Fig. 3).
Discussion
The results of the present study demonstrated that
breast cancer tissues exhibited higher levels of Cbx2 expression
compared with normal tissues (53.85 vs. 11.11%). Aberrant
expression of Cbx2 at the mRNA level has been observed in colon,
breast, stomach and lung cancer (27). In the present study, Cbx2 protein
expression was associated with certain clinicopathological features
in breast cancer patients. High Cbx2 expression was found to
associate with a large tumor size (P<0.001), and this finding
was consistent with the results of a previous oral squamous cell
carcinoma study (28). In addition,
Cbx2 was significantly associated with lymph node metastasis
(P=0.008) and a positive HER-2 status (P=0.048). A positive
association was also identified between Cbx2 expression and TNM
stage (P<0.001). At present, the function of Cbx2 in
tumorigenesis remains unclear, however emerging evidence has
indicated that the Cbx2 protein exhibits a critical role in cancer
initiation and progression (29,30).
Cbx2 is a primary member of the Cbx protein family,
and is a component of the PRC1 complex that regulates chromatin.
PRC1 exhibits enzymatic activity to modify histones and repress the
transcription of target genes (31,32). The
ability of PRC1 to promote proliferation may be associated with PcG
activity in cancer (33). The Cbx2
protein is a major component involved in the recruitment of PRC1
proteins to mitotic chromosomes (34). Cbx2 also directly regulates the
expression of the cyclin-dependent kinase inhibitor, p21, and
dominantly controls the expression of the INK4A/ARF locus, which is
extremely important for human hematopoietic cell proliferation
(35). The overexpression of Cbx2
results in the differentiation and exhaustion of hematopoietic stem
cells (16). Previous studies
regarding the mechanism of Cbx2 in regulating hematological stem
cell differentiation have been conducted, however, few studies have
investigated the function of Cbx2 in solid tumors (16,36). The
present study provides a basis for future functional studies to
identify molecular mechanisms by which Cbx2 may promote tumor
initiation and progression.
The current study indicated that Cbx2 expression
associates with the prognosis of breast cancer patients. At the
final follow-up, the mortality rates in the high and low Cbx2
expression groups were 18.64 and 10.93%, respectively. The mean
survival time in the high Cbx2 expression group (74.29 months) was
significantly shorter than that in the low expression group (77.37
months). High Cbx2 expression was also significantly associated
with poor OS (HR, 1.826; 95% CI, 1.069–3.116; P=0.027). Considering
the poor prognosis of patients with high Cbx2 expression, we
hypothesize that Cbx2 may present an important predictor of breast
cancer prognosis. By integrating multiple platforms on several
biological levels, Clermont et al (27) demonstrated that an increased Cbx2 copy
number associates with increased Cbx2 expression, which is
significantly associated with poor OS. Due to the heterogeneity
observed in the progression and outcome of breast cancer, the
identification of more predictive biomarkers is required to guide
clinical treatment.
Taxol is a microtubule-stabilizing drug approved by
the Food and Drug Administration for the treatment of breast cancer
(37). A previous study revealed that
breast cancer patients that received nab-paclitaxel neoadjuvant
chemotherapy exhibited a pathological complete response rate of
48.1% (38). Despite the high
response rate, certain patients exhibit low or no response to
Taxol. Thus, additional effective biomarkers are required to
identify which patients would benefit from Taxol therapy. In the
current study, the OS time of patients treated with Taxol was
significantly lower than that of patients who did not receive Taxol
treatment in the high Cbx2 expression group. However, no
significant difference in OS time was identified between patients
treated with or without Taxol in the low Cbx2 expression group. The
association between Cbx2 expression and survival of breast cancer
patients treated with fluorouracil was also analyzed in the present
study. No significant difference was identified between the OS time
of patients with and without fluorouracil treatments in the high or
low Cbx2 expression groups (data not provided). Thus, Cbx2 may
present a specific biomarker for Taxol resistance in breast cancer
patients. Numerous studies have been conducted to identify marker
resistance or sensitivity to Taxol. These studies identified
various candidates, including adenosine triposphate-binding
cassette subfamily C member10, microRNA and solute carrier genes
(39–41). However, these studies did not identify
a validated biomarker to predict which patients would benefit from
Taxol therapy. Cbx2 may present a promising indicator for
predicting the use of Taxol in breast cancer chemotherapy. However,
further clinical studies are required to verify these results.
The present study demonstrated the association
between Cbx2 expression and breast cancer. However, certain points
require further study. Firstly, dynamic localization of the
nuclear-cytoplasmic and/or sub-nuclear distribution of members of
the Cbx family occurs during the maternal-to-embryonic transition
(42). The present study revealed
that Cbx2 was predominantly expressed in the cytoplasm. Nuclear
proteins under normal conditions are frequently overexpressed in
the cytoplasm in various human cancers, including hepatocellular
carcinoma, melanoma, papillary thyroid carcinoma and ductal breast
carcinoma (43,44). Secondly, tumorigenesis is a dynamic
evolutionary process that promotes genetic heterogeneity and
produces a complex combination of random and nonrandom aberrations.
The current study investigated Cbx2 expression at the protein level
and thus, results must be confirmed by integrating multiple
platforms on several biological levels (DNA-RNA-protein). Thirdly,
all samples in the current study were recruited from a single
hospital. Due to the heterogeneity of breast cancer, future studies
which include individuals of different ethnicities in the patient
cohort are required to validate the results of the present study.
Furthermore, studies which aim to elucidate the molecular
mechanisms underlying Cbx2 protein expression and its function in
tumorigenesis are required.
In conclusion, in the present study Cbx2 protein
expression levels were inversely associated with prognosis in
breast cancer patients. Cbx2 expression was associated with
clinical features, including positive lymph node metastasis status,
large tumor size and positive HER-2 status. Therefore, Cbx2 may
present a novel biomarker for the selection of an appropriate
chemotherapy regimen for breast cancer patients.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ali AM, Provenzano E, Bartlett JM, Abraham
J, Driver K, Munro AF, Twelves C, Poole CJ, Hiller L, Dunn JA, et
al: Prognosis of early breast cancer by immunohistochemistry
defined intrinsic sub-types in patients treated with adjuvant
chemotherapy in the NEAT/BR9601 trial. Int J Cancer. 133:1470–1478.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu R, Lv QL, Yu J, Hu L, Zhang LH, Cheng
Y and Zhou HH: Correlating transcriptional networks with
pathological complete response following neoadjuvant chemotherapy
for breast cancer. Breast Cancer Res Treat. 151:607–618. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Krishnan P, Ghosh S, Wang B, Li D,
Narasimhan A, Berendt R, Graham K, Mackey JR, Kovalchuk O and
Damaraju S: Next generation sequencing profiling identifies
miR-574-3p and miR-660-5p as potential novel prognostic markers for
breast cancer. BMC Genomics. 16:7352015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boccardo F, Rubagotti A, Nuzzo PV,
Argellati F, Savarino G, Romano P, Damonte G, Rocco M and Profumo
A: Matrix-assisted laser desorption/ionisation (MALDI) TOF analysis
identifies serum angiotensin II concentrations as a strong
predictor of all-cause and breast cancer (BCa)-specific mortality
following breast surgery. Int J Cancer. 137:2394–2402. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sparmann A and van Lohuizen M: Polycomb
silencers control cell fate, development and cancer. Nat Rev
Cancer. 6:846–856. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shao Z, Raible F, Mollaaghababa R, Guyon
JR, Wu CT, Bender W and Kingston RE: Stabilization of chromatin
structure by PRC1, a Polycomb complex. Cell. 98:37–46. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Levine SS, Weiss A, ErdjumentBromage H,
Shao Z, Tempst P and Kingston RE: The core of the polycomb
repressive complex is compositionally and functionally conserved in
flies and humans. Mol Cell Biol. 22:6070–6078. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuzmichev A, Nishioka K, ErdjumentBromage
H, Tempst P and Reinberg D: Histone methyltransferase activity
associated with a human multiprotein complex containing the
Enhancer of Zeste protein. Genes Dev. 16:2893–2905. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vincenz C and Kerppola TK: Different
polycomb group CBX family proteins associate with distinct regions
of chromatin using nonhomologous protein sequences. Proc Natl Acad
Sci USA. 105:16572–16577. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shinjo K, Yamashita Y, Yamamoto E,
Akatsuka S, Uno N, Kamiya A, Niimi K, Sakaguchi Y, Nagasaka T,
Takahashi T, et al: Expression of chromobox homolog 7 (CBX7) is
associated with poor prognosis in ovarian clear cell adenocarcinoma
via TRAIL-induced apoptotic pathway regulation. Int J Cancer.
135:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Scott CL, Gil J, Hernando E,
TeruyaFeldstein J, Narita M, Martínez D, Visakorpi T, Mu D,
Cordon-Cardo C, Peters G, et al: Role of the chromobox protein CBX7
in lymphomagenesis. Proc Natl Acad Sci USA. 104:5389–5394. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang XW, Zhang L, Qin W, Yao XH, Zheng
LZ, Liu X, Li J and Guo WJ: Oncogenic role of the chromobox protein
CBX7 in gastric cancer. J Exp Clin Cancer Res. 29:1142010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei
Z, Yin QQ, Ma LN, Zhou AW, Wang LS, et al: Cbx4 governs HIF-1α to
potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3
ligase activity. Cancer Cell. 25:118–131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Klauke K, Radulović V, Broekhuis M,
Weersing E, Zwart E, Olthof S, Ritsema M, Bruggeman S, Wu X, Helin
K, et al: Polycomb Cbx family members mediate the balance between
haematopoietic stem cell self-renewal and differentiation. Nat Cell
Biol. 15:353–362. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Parris TZ, Danielsson A, Nemes S, Kovács
A, Delle U, Fallenius G, Möllerström E, Karlsson P and Helou K:
Clinical implications of gene dosage and gene expression patterns
in diploid breast carcinoma. Clin Cancer Res. 16:3860–3874. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH
and van de Vijver MJ: WHO Classification of Tumours of the Breast.
4th. IARC, WHO; 2012
|
|
19
|
Nishimukai A, Yagi T, Yanai A, Miyagawa Y,
Enomoto Y, Murase K, Imamura M, Takatsuka Y, Sakita I, Hatada T and
Miyoshi Y: High Ki-67 expression and low progesterone receptor
expression could independently lead to a worse prognosis for
postmenopausal patients with estrogen receptor-positive and
HER2-negative breast cancer. Clin Breast Cancer. 15:204–211. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al: American Society of Clinical Oncology;
College of American Pathologists: Recommendations for human
epidermal growth factor receptor 2 testing in breast cancer:
American Society of Clinical Oncology/College of American
Pathologists clinical practice guideline update. Arch Pathol Lab
Med. 138:241–256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tubbs RR, Hicks DG, Cook J, DownsKelly E,
Pettay J, Hartke MB, Hood L, Neelon R, Myles J, Budd GT, et al:
Fluorescence in situ hybridization (FISH) as primary methodology
for the assessment of HER2 Status in adenocarcinoma of the breast:
A single institution experience. Diagn Mol Pathol. 16:207–210.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Falato C, Lorent J, Tani E, Karlsson E,
Wright PK, Bergh J and Foukakis T: Ki67 measured in metastatic
tissue and prognosis in patients with advanced breast cancer.
Breast Cancer Res Treat. 147:407–414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Millar EK, Graham PH, McNeil CM, Browne L,
O'Toole SA, Boulghourjian A, Kearsley JH, Papadatos G, Delaney G,
Fox C, et al: Prediction of outcome of early ER+ breast cancer is
improved using a biomarker panel, which includes Ki-67 and p53. Br
J Cancer. 105:272–280. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamashita H, Toyama T, Nishio M, Ando Y,
Hamaguchi M, Zhang Z, Kobayashi S, Fujii Y and Iwase H: p53 protein
accumulation predicts resistance to endocrine therapy and decreased
post-relapse survival in metastatic breast cancer. Breast Cancer
Res. 8:R482006. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Remmele W and Stegner HE: Recommendation
for uniform definition of an immunoreactive score (IRS) for
immunohistochemical estrogen receptor detection (ER-ICA) in breast
cancer tissue. Pathologe. 8:138–140. 1987.(In German). PubMed/NCBI
|
|
26
|
Remmele W and Schicketanz KH:
Immunohistochemical determination of estrogen and progesterone
receptor content in human breast cancer. Computer-assisted image
analysis (QIC score) vs subjective grading (IRS). Pathol Res Pract.
189:862–866. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Clermont PL, Sun L, Crea F, Thu KL, Zhang
A, Parolia A, Lam WL and Helgason CD: Genotranscriptomic
meta-analysis of the Polycomb gene CBX2 in human cancers: Initial
evidence of an oncogenic role. Br J Cancer. 111:1663–1672. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Parris TZ, Aziz L, Kovács A, Hajizadeh S,
Nemes S, Semaan M, Chen CY, Karlsson P and Helou K: Clinical
relevance of breast cancer-related genes as potential biomarkers
for oral squamous cell carcinoma. BMC Cancer. 14:3242014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Satijn DP, Olson DJ, van der Vlag J, Hamer
KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R
and Otte AP: Interference with the expression of a novel human
polycomb protein, hPc2, results in cellular transformation and
apoptosis. Mol Cell Biol. 17:6076–6086. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang Y, Malouf GG, Zhang J, Zheng X, Chen
Y, Thompson EJ, Weinstein JN, Yuan Y, Spano JP, Broaddus R, et al:
Long non-coding RNA profiling links subgroup classification of
endometrioid endometrial carcinomas with trithorax and polycomb
complex aberrations. Oncotarget. 6:39865–39876. 2015.PubMed/NCBI
|
|
31
|
Kerppola TK: Polycomb group complexes-many
combinations, many functions. Trends Cell Biol. 19:692–704. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Croce L and Helin K: Transcriptional
regulation by Polycomb group proteins. Nat Struct Mol Biol.
20:1147–1155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Piunti A, Rossi A, Cerutti A, Albert M,
Jammula S, Scelfo A, Cedrone L, Fragola G, Olsson L, Koseki H, et
al: Polycomb proteins control proliferation and transformation
independently of cell cycle checkpoints by regulating DNA
replication. Nat Commun. 5:36492014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhen CY, Duc HN, Kokotovic M, Phiel CJ and
Ren X: Cbx2 stably associates with mitotic chromosomes via a PRC2-
or PRC1-independent mechanism and is needed for recruiting PRC1
complex to mitotic chromosomes. Mol Biol Cell. 25:3726–3739. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
van den Boom V, RozenveldGeugien M,
Bonardi F, Malanga D, van Gosliga D, Heijink AM, Viglietto G,
Morrone G, Fusetti F, Vellenga E and Schuringa JJ: Nonredundant and
locus-specific gene repression functions of PRC1 paralog family
members in human hematopoietic stem/progenitor cells. Blood.
121:2452–2461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van den Boom V, RozenveldGeugien M,
Bonardi F, Malanga D, van Gosliga D, Heijink AM, Viglietto G,
Morrone G, Fusetti F, Vellenga E and Schuringa JJ: Nonredundant and
locus-specific gene repression functions of PRC1 paralog family
members in human hematopoietic stem/progenitor cells. Blood.
121:2452–2561. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weaver BA: How Taxol/paclitaxel kills
cancer cells. Mol Biol Cell. 25:2677–2681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zelnak AB, Nikolinakos P, Srinivasiah J,
Jonas W, Pippas A, Liu Y, Li X, Torres M and O'Regan RM: Georgia
Center for Oncology Research and Education: High pathologic
complete response in Her2-positive, early-stage breast cancer to a
novel nonanthracycline neoadjuvant chemotherapy. Clin Breast
Cancer. 15:31–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kathawala RJ, Sodani K, Chen K, Patel A,
Abuznait AH, Anreddy N, Sun YL, Kaddoumi A, Ashby CR Jr and Chen
ZS: Masitinib antagonizes ATP-binding cassette subfamily C member
10-mediated paclitaxel resistance: A preclinical study. Mol Cancer
Ther. 13:714–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peng X, Cao P, He D, Han S, Zhou J, Tan G,
Li W, Yu F, Yu J, Li Z and Cao K: MiR-634 sensitizes nasopharyngeal
carcinoma cells to paclitaxel and inhibits cell growth both in
vitro and in vivo. Int J Clin Exp Pathol. 7:6784–6791.
2014.PubMed/NCBI
|
|
41
|
Russell P, Hennessy BT, Li J, Carey MS,
Bast RC, Freeman T and Venkitaraman AR: Cyclin G1 regulates the
outcome of taxane-induced mitotic checkpoint arrest. Oncogene.
31:2450–2460. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ruddock-D'Cruz NT, Prashadkumar S, Wilson
KJ, Heffernan C, Cooney MA, French AJ, Jans DA, Verma PJ and
Holland MK: Dynamic changes in localization of Chromobox (Cbx)
family members during the maternal to embryonic transition. Mol
Reprod Dev. 75:477–488. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huynh H: Overexpression of tumour
suppressor retinoblastoma 2 protein (pRb2/p130) in hepatocellular
carcinoma. Carcinogenesis. 25:1485–1494. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Reno EM, Haughian JM, Dimitrova IK,
Jackson TA, Shroyer KR and Bradford AP: Analysis of protein kinase
C delta (PKC delta) expression in endometrial tumors. Hum Pathol.
39:21–29. 2008. View Article : Google Scholar : PubMed/NCBI
|