Introduction
Gastric cancer is a common malignancy affecting the
gastrointestinal system, which accounts for 8% of all types of
malignant cancer. Although the incidence of gastric cancer has been
declining worldwide, it remains high in several countries,
particularly in China (1). The
development and progression of gastric cancer are associated with
multiple genes and numerous transforming steps. Investigating
genetic and other risk factors associated with the occurrence and
development of gastric cancer, and their correlation with
prognosis, is critical to provide theoretical support for improving
the diagnosis, prognosis and treatment of gastric cancer (2). MicroRNAs (miRs) are small non-coding RNA
molecules of ~22 nucleotides, which regulate gene expression at the
post-transcriptional level. The regulatory capacity of miRs
includes almost all major cellular activities, including cell
proliferation, differentiation and apoptosis. In previous years, a
number of studies have shown that microRNAs can have oncogene and
tumor suppressor functions, thereby being involved in tumor
development and progression (3–5). A
previous study showed that miR-146a was associated with gastric
cancer metastasis through modulating Wiskott-Aldrich syndrome
protein family member 2 (6). Other
studies have shown that the expression of miR-146a is associated
with tumor cell proliferation and apoptosis in gastric cancer
(7,8).
miR-146a has direct effects on tumor metastasis and prognosis by
affecting epidermal growth factor receptor (EGFR) (8). The expression of LIN52 in
gastrointestinal tumors affects drug sensitivity and enhances the
effect of imatinib-induced apoptosis in tumor cells (9). In addition, according to the predictions
of TargetScan, miR-146a is a target gene regulated by LIN52
(10). The present study further
analyzed the effect of miR-146a on the treatment of gastric cancer.
The expression of miR-146a was analyzed in 93 clinical cases of
advanced gastric cancer using reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis, and its expression
was correlated with the clinical characteristics of advanced
gastric cancer and the prognosis of patients.
Materials and methods
Patients and sampling
A total of 93 patients with advanced gastric cancer
(57 men and 36 women; median age, 61 years), who underwent surgical
treatment between June 2009 and January 2011 in Henan Provincial
People's Hospital (Henan, China) were enrolled in the present
study. All patients received an oxaliplatin-based chemotherapeutic
regimen following surgical treatment, and the efficacy was
evaluated in accordance with the Response Evaluation Criteria in
Solid Tumors (RESIST) (11) The
histopathological analysis of resected tissues was based on the
tumor-node-metastasis (TNM) staging criteria of the World Health
Organization (11). From each
patient, non-tumor tissues were collected 7 cm adjacent to the
tumor lesion as an internal control. The histological sections of
each resected specimen were classified by pathologists. All
patients signed written informed consent prior to their involvement
in the study. The experimental protocol of the present study was
approved by the Ethics Committee of Henan Provincial People's
Hospital.
Reagents
Mouse anti-human LIN52 monoclonal antibody was
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
The streptavidin-peroxidase (SP) immunohistochemical staining kit
and 3,3′-diaminobenzidine (DAB) peroxidase substrate kit were
purchased from Fuzhou Maxim Biotech, Inc. (Fujian, China). The
experimental procedures for the immunohistochemistry were according
to the protocol of the SP staining kit manufacturer.
Phosphate-buffered saline (PBS) with Tween detergent was used to
replace primary antibody in the negative control for the
immunohistochemistry assays. TRIzol reagent was purchased from
Thermo Fisher Scientific, Inc. (Waltham, MA, USA). The reverse
transcription kit (K1622 MBI) was purchased from Fermentas; Thermo
Fisher Scientific, Inc. SYBR® Green PCR Master Mix was
purchased from Takara Bio, Inc. (Otsu, Japan). The Stratagene
Mx3005P RT-qPCR instrument was purchased from Agilent Technologies,
Inc. (Santa Clara, CA, USA). Other conventional reagents for
molecular biology were purchased from Sigma-Aldrich (Merck
Millipore, Darmstadt, Germany).
Immunohistochemistry
Following conventional tissue processing,
paraffin-embedding and sectioning, each tissue section was dewaxed
and hydrated in water, followed by incubation with 3% hydrogen
peroxide at room temperature for 15 min and high pressure antigen
retrieval for 2 min. Following cooling of the sections to room
temperature, each section was blocked in normal goat serum (Fuzhou
Maxim Biotech, Inc.) at room temperature for 10 min, and incubated
with LIN52 antibody (dilution, 1:300) at 4°C overnight. Each tissue
section was then incubated with biotinylated rabbit anti-mouse
secondary antibody (Santa Cruz Biotechnology, Inc.) at 37°C for 10
min. Following washing of the tissue sections with PBS, DAB
solution was used for color development, followed by repeated
washing under tap water and hematoxylin counterstaining. Excess dye
from the tissue sections was removed using hydrochloric acid. All
tissue sections were dehydrated in graded ethanol solutions and
cleared in xylene solution prior to mounting on glass slides using
neutral gum. A light microscope was used to visualize staining.
RT-qPCR analysis
Following collection of the resected tissue and snap
freezing in liquid nitrogen, each tissue specimen was crushed into
a powder, followed by total RNA extraction. TRIizol reagent (1 ml)
was added to the tissue powder to lyse the cells for 10 min,
followed by transfer of the supernatant to a microcentrifuge tube.
Chloroform solution (400 µl) was added to the supernatant, vortexed
and subjected to 12,000 g centrifugation at 4°C for 15 min. The
supernatant (200 µl) was then transferred to a new RNase-free
microcentrifuge tube and mixed with equal quantities of isopropanol
by inverting the tube, followed by incubation for 10 min and 12,000
g centrifugation at 4°C for 10 min. The supernatant was discarded,
followed by the addition of 1 ml 70% ethanol to the pellet and
mixing by gentle inversion of the microcentrifuge tube. The mixture
was centrifuged at 12,000 g at 4°C for 10 min, following which the
ethanol solution was discarded and the pellet was air-dried. The
pellet was then dissolved in diethyl pyrocarbonate-treated
distilled water. The total RNA concentration of each sample was
determined using an e-Spect ultra-small spectrophotometer (Malcom
Co., Ltd., Tokyo, Japan). The RNA quality of each sample was
determined based on the optical density (OD) 260/280 ratio, ranging
between 1.8 and 2.0. For cDNA synthesis, 20 µl reverse
transcriptase solution from the kit was added to 0.1 µg RNA
template, miR-146a reverse primer and U6 primer for reverse
transcription into cDNA. U6 was the internal reference for analysis
of the expression of miR-146a. Details of the primers are listed in
Table I. The primers and RNA template
were incubated at 65°C for 5 min, followed by cooling on ice,
incubation with the reverse transctiption reaction mixture and
dNTPs at 42°C for 60 min, and termination of the reaction at 70°C
for 5 min. The total volume of master mix for the qPCR analysis was
20 µl, containing 1 µl of cDNA (final concentration 5 ng), 12.5 µl
of 2X SYBR Green I Master mix, 0.5 µmol/l miR-146a/U6 specific
forward primers and 0.5 µmol/l miR-146a/U6 reverse primers. Each
sample had three replicates. The qPCR conditions were as follows:
95°C for 7 min, followed by 40 cycles of denaturation at 95°C for
15 sec, annealing at 60°C for 25 sec and elongation at 72°C for 25
sec. The quantification cycle (Cq) value of the U6 reaction
obtained from the qPCR analysis was used to calculate the Cq value
representing the relative expression of miR-146a. qPCR was
activated at 95°C for 5 min, followed by 45 cycles of denaturation
at 95°C for 15 sec, annealing at 62°C for 30 sec and elongation at
72°C for 20 sec. The Cq value, standard and melting curves were
automatically generated by the RT-qPCR instrument.
 | Table I.miR-146a and U6 primer sequences for
reverse transcription-quantitative polymerase chain reaction
analysis. |
Table I.
miR-146a and U6 primer sequences for
reverse transcription-quantitative polymerase chain reaction
analysis.
| Primer | Sequence |
|---|
| miR-146a |
|
| RT |
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACCCATG-3′ |
|
Forward |
5′-TGGTGTCGTGGAGTCG-3′ |
|
Reverse |
5′-ACACTCCAGCTGGGTGAGAACTGAATTCCATGGGTT-3′ |
| U6 |
|
|
Forward |
5′-CTCGCTTCGGCAGCACA-3′ |
| RT and
reverse |
5′-AACGCTTCACGAATTTGCGT-3′ |
Statistical analysis
SPSS 13.0 software was used for the non-parametric
rank sum test. Relative expression ≥2.0 was defined as high
expression and relative expression <2.0 was defined as low
expression following Kaplan-Meier survival analysis. The Cox
regression model was based on the forward method of conditional
parameter estimates. P<0.05 was considered to indicate a
statistically significant difference.
Results
Positive protein expression of
LIN52
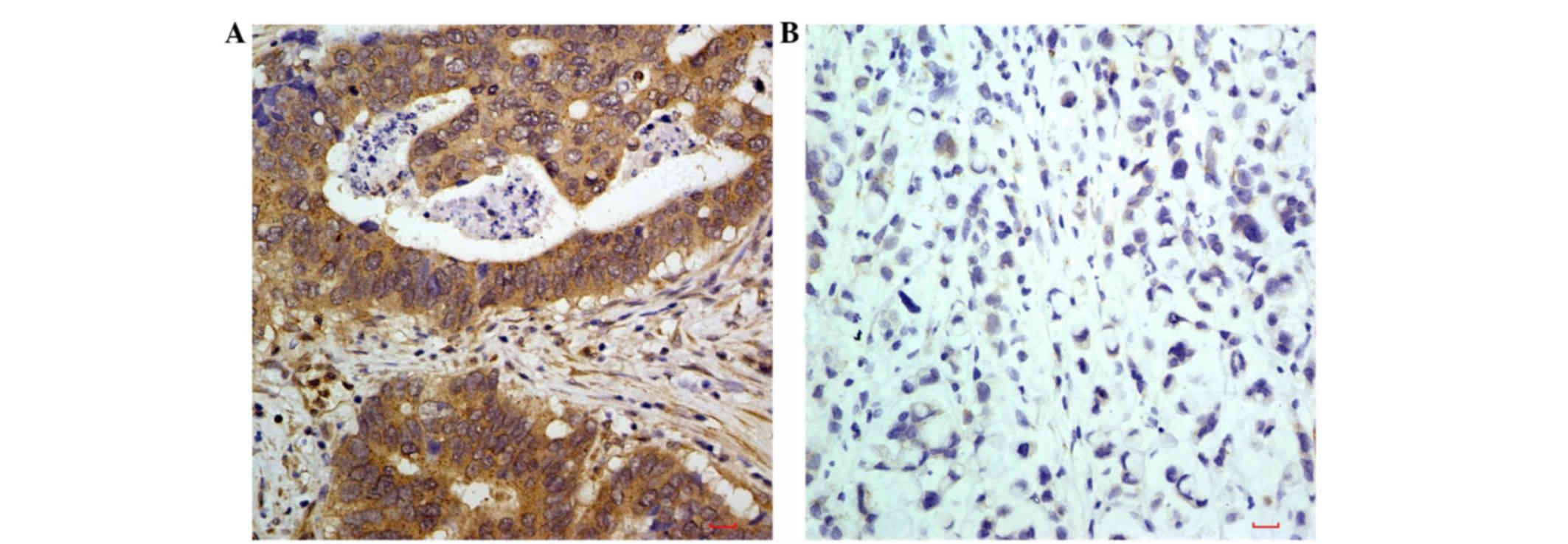
As shown in Fig. 1A,
positive staining of the LIN52 protein, identified as brownish
yellow granules, was distributed predominantly in the cytoplasm of
the advanced gastric cancer tissues. In the normal tissues adjacent
to the tumor, the expression of LIN52 was low (Fig. 1B).
Correlation between the expression of
miR-146a in advanced gastric cancer tissues and clinicopathological
parameters
The expression of miR-146a in the advanced gastric
cancer tissue was significantly correlated with the clinical TNM
staging of the patients (P<0.05). The expression of miR-146a in
stage III gastric cancer tissues was significantly higher, compared
with that in stage IV gastric cancer tissues (P<0.05). Patients
with lymph node metastasis had lower expression levels of miR-146a,
compared with patients without lymph node metastasis. However, no
significant correlation was found between the expression of
miR-146a in advanced gastric cancer tissue and other
clinicopathological factors, including gender, age and tumor
differentiation (P>0.05; Table
II).
 | Table II.Expression of miR-146a and LIN52 in 93
patients with gastric cancer and their correlation with
clinicopathological parameters. |
Table II.
Expression of miR-146a and LIN52 in 93
patients with gastric cancer and their correlation with
clinicopathological parameters.
|
|
| miR-146a, n
(%) | LIN52, n (%) |
|---|
|
|
|
|
|
|---|
| Parameter | n | Low | High |
P-valuea | Low | High |
P-valuea |
|---|
| Gender |
|
|
| 0.091 |
|
| 0.077 |
|
Male | 57 | 34 (59.6) | 23 (40.4) |
| 29 (53.7) | 25 (46.3) |
|
|
Female | 36 | 15 (41.7) | 21 (58.3) |
| 28 (71.8) | 11 (28.2) |
|
| Age (years) |
|
|
| 0.350 |
|
| 0.484 |
|
<56 | 32 | 19 (59.4) | 13 (40.6) |
| 18 (56.3) | 14 (43.8) |
|
|
≥56 | 61 | 30 (49.2) | 31 (50.8) |
| 39 (63.9) | 22 (36.1) |
|
| Grade |
|
|
| 0.295 |
|
| 0.519 |
| Poorly
differentiated | 18 | 13 (72.2) | 5
(27.8) |
| 5
(27.8) | 13 (72.2) |
|
|
Moderately differentiated | 33 | 17 (51.5) | 16 (48.5) |
| 15 (45.5) | 18 (54.5) |
|
| Well
differentiated | 22 | 10 (45.5) | 12 (54.5) |
| 11 (50.0) | 11 (50.0) |
|
|
Signet-ring cell
carcinoma | 20 | 9
(45.0) | 11 (55.5) |
| 8
(40.0) | 12 (60.0) |
|
| Lymph node
metastasis |
|
|
| 0.033 |
|
| 0.754 |
|
Present | 76 | 44 (57.9) | 32 (42.1) |
| 39 (59.1) | 27 (40.9) |
|
| Not
present | 17 | 5
(29.4) | 12 (70.6) |
| 15 (55.6) | 12 (44.4) |
|
| Clinical stage |
|
|
| 0.011 |
|
| 0.051 |
|
III | 42 | 16 (38.1) | 26 (61.9) |
| 29 (69.0) | 13 (31.0) |
|
| IV | 51 | 33 (64.7) | 18 (35.3) |
| 25 (49.0) | 26 (51.0) |
|
Correlation between the expression of
miR-146a and patient survival rates
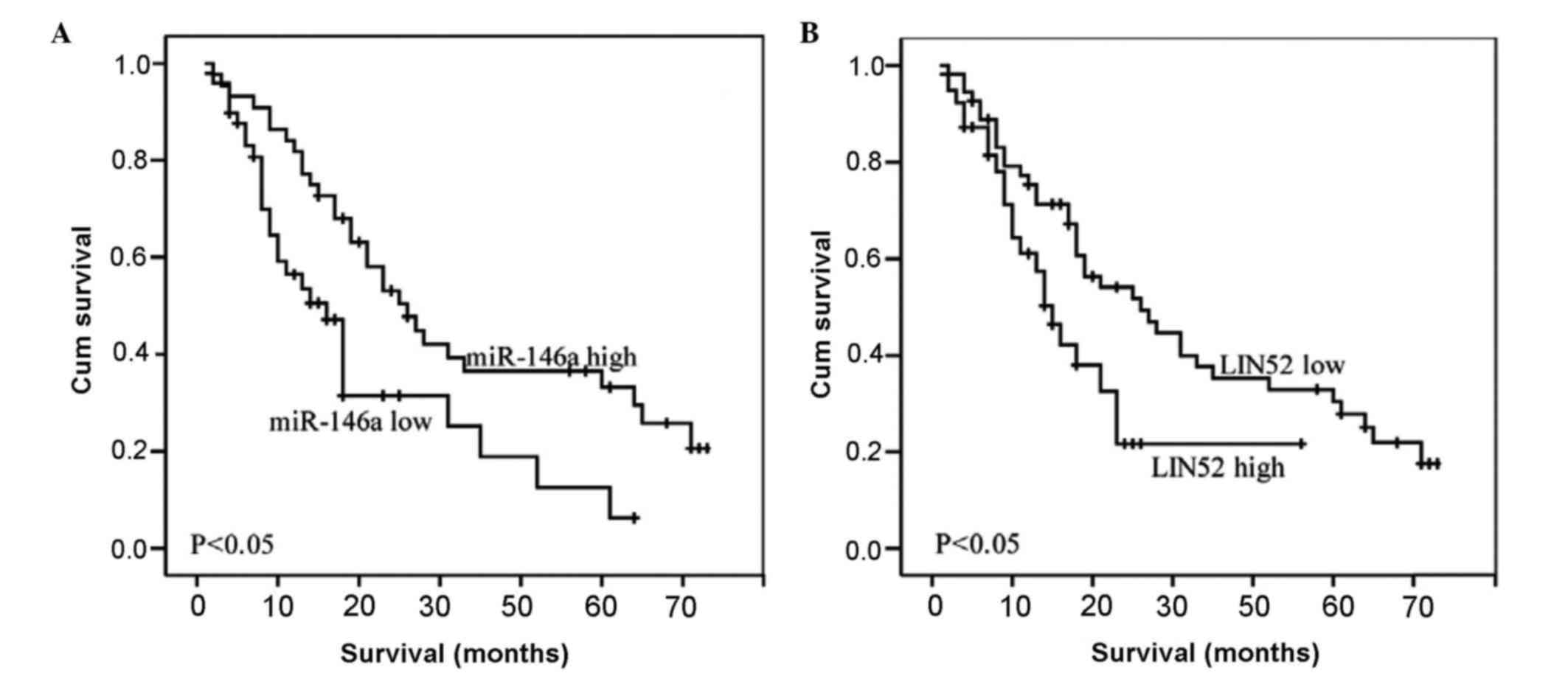
The results of the Kaplan-Meier survival analysis
showed that the expression of miR-146a had a significant effect on
the survival rates of patients with advanced gastric cancer.
Patients with high expression levels of miR-146a had significantly
higher survival rates, compared with patients with low expression
levels of miR-146a (P<0.05; Fig.
2).
Correlation between the expression
levels of miR146a and LIN52 in advanced gastric cancer
The present study further analyzed the correlation
between the expression levels of miR146a and LIN52. Among the 44
specimens with a high expression of miR146a, only 8 (18.2%) showed
LIN52 immunoreactivity (P<0.001; Table III).
 | Table III.Correlation between the expression of
miR146a and LIN52. |
Table III.
Correlation between the expression of
miR146a and LIN52.
|
|
| LIN52 |
|
|---|
|
|
|
|
|
|---|
| miR146a | n | Low, n (%) | High, n (%) |
P-valuea |
|---|
| Low | 49 | 21 (42.9) | 28 (57.1) | <0.001 |
| High | 44 | 36 (81.8) | 8
(18.2) |
Cox regression analysis of prognostic
factors in patients with gastric cancer
The clinical prognostic factors of patients with
advanced gastric cancer were used as dependent variables and their
relevant effects were used as independent variables for Cox
regression analysis. As shown in Table
IV, TNM staging, lymph node metastasis and the expression of
miR-146a were independent risk factors for the prognosis of
patients with advanced gastric cancer.
 | Table IV.Cox regression analysis of prognostic
factors in patients with gastric cancer. |
Table IV.
Cox regression analysis of prognostic
factors in patients with gastric cancer.
|
| 95.0% CI for
HR |
|---|
|
|
|
|---|
| Factor | Regression
coefficient | SE | χ2 |
P-valuea | HRa | Lower | Upper |
|---|
| TNM staging |
0.732 | 0.345 | 4.489 | 0.034 | 2.079 | 1.056 | 4.09 |
| Lymph node
metastasis | −2.512 | 1.088 | 5.325 | 0.021 | 0.0081 | 0.010 | 0.685 |
| microRNA-146a
expression | −2.048 | 0.785 | 6.806 | 0.009 | 0.129 | 0.028 | 0.601 |
Expression of miR-146a and
chemotherapeutic sensitivity
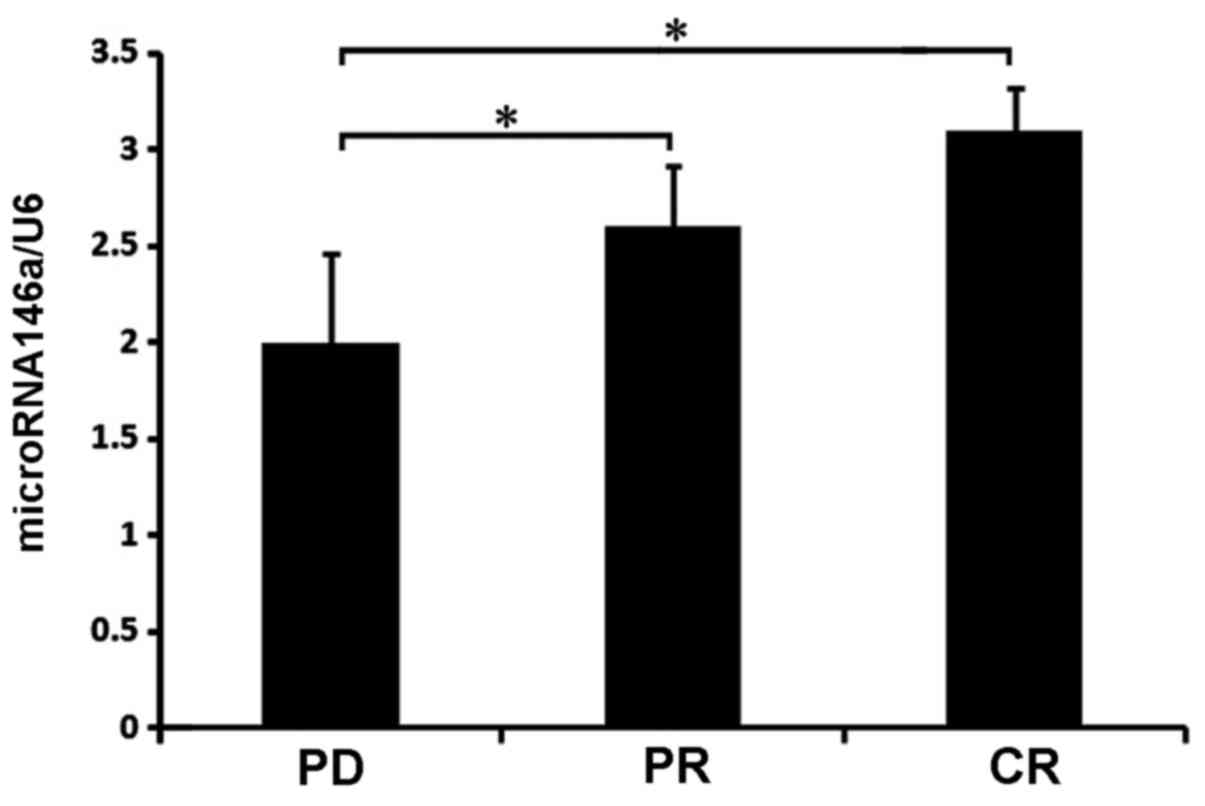
The present study also assessed the association
between the expression of miR-146a and chemotherapeutic sensitivity
in patients with advanced gastric cancer. The analysis showed that
patients with advanced gastric cancer tissues expressing a low
level of miR-146a had a poor prognosis (Fig. 3). The present study evaluated the
efficacy of chemotherapy in patients with advanced gastric cancer
according to the RESIST standard and divided the patients into
three groups: Complete remission (CR), partial remission (PR) and
disease progression (PD). The characteristics of the expression of
miR-146a were analyzed in the three groups. The expression of
miR-146a in the CR group was the highest, with significant
differences between the PR and PD groups, and the PR and PD groups
(P<0.05), but not between the PR and CR groups (P>0.05).
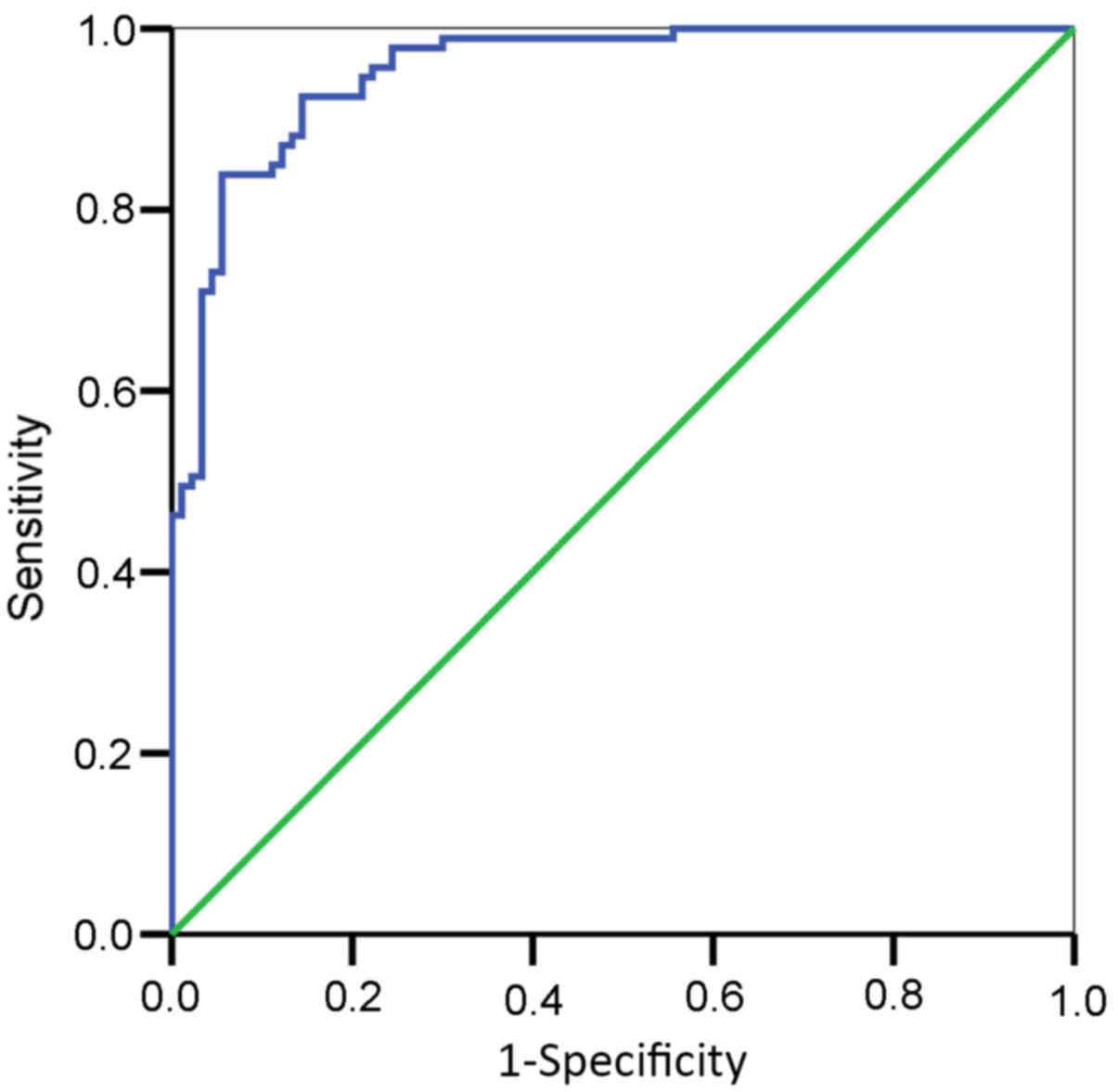
Evaluation of diagnostic specificity
and sensitivity of miR-146a in patients with advanced gastric
cancer using ROC curve regression analysis
The area under the ROC curve of miR-146a in the
advanced gastric cancer tissues and corresponding adjacent
non-tumor tissue was 0.760 [5% confidence interval
(CI)=0.589–0.942; P=0.016], suggesting that miR-146a was
significant for distinguishing between advanced gastric cancer
tissue and normal tissue (P<0.05). Lower expression levels of
miR-146a increased the likelihood of advanced gastric cancer. These
results indicated the suitability of using miR-146a as an adjuvant
diagnostic marker for advanced gastric cancer. In the present
study, the relative expression of miR-146a was 19.75, which was
used as the evaluation threshold for the diagnosis of advanced
gastric cancer, with 94.1% sensitivity and 61.5% specificity
(Fig. 4).
Discussion
Gastric cancer is one of the most common types of
cancer in China, with a high incidence rate and insidious early
symptoms (12). At the time of
diagnosis, patients with gastric cancer are often at moderate
and/or advanced stages of the disease, with poor prognosis
(11). Therefore, the degree of
malignancy and the mortality rates of patients with gastric cancer
are relatively high. At present, there is no method for the
effective early diagnosis of gastric cancer and, if diagnosed with
an advanced stage of disease, radical surgery is not an option for
patients. Advances in molecular targeted therapy and chemotherapies
have not significantly improved the median survival rates of
patients with advanced gastric cancer (13). Although α-fetoprotein,
carcinoembryonic antigen, CA125 and CA199 enhance the early
diagnostic sensitivity of gastric cancer, their diagnostic
specificities as markers of gastric cancer are low (14). It is necessary to investigate novel
diagnostic markers for gastric cancer to provide guidance for
cancer diagnosis and treatment (15).
To address this problem, studies are focusing on the use of miRs in
gastric cancer as potential biomarkers (16). Evaluation of the expression of miR in
tumor tissue and patient serum is important in the early diagnosis
and prognosis of gastric cancer (17). In the present study, the expression of
miR-146a in gastric cancer tissue was correlated with the early
diagnosis of gastric cancer and the prognosis of patients, and
predictive analysis on these parameters was performed.
Through binding with the mRNA 3′-untranslated region
(3′UTR), a single miR may have regulatory effects against various
mRNAs. Typically, there is differential expression of miRs in tumor
tissues, compared with normal tissues. miRs usually have specific
and stable expression in tissues, therefore, miRs offer potential
as prognostic markers (18,19). Binding between miRs and the 3′UTR of
mRNA suppresses mRNA transcription and affects the expression of
oncogenes and tumor suppressor genes, thereby promoting or
inhibiting tumor occurrence, development and metastasis. A previous
study showed that miRs can be used as potential targets in
anticancer therapy (20). Another
prevopis study showed that miRs are associated with tumor
metastasis, tolerance of anticancer therapy and tumor progression
(21,22). In addition, miRs have only ~20
nucleotide bases, which facilitates its stable expression in
tissues and its detection (23).
Therefore, miRs have received increasing attention in tumor
diagnosis, treatment evaluation and determining the prognosis of
cancer (24,25).
Although the biological functions of miRs remain to
be fully elucidated, the expression of miR in normal tissue,
compared with tumor tissue, is often apparent. To date, increasing
applications of miRs as tissue-specific markers have become
available for the analysis of primary tumor metastasis (26,27). The
present study showed that the relative expression of miR-146a in
gastric cancer tissue was significantly lower, compared with that
in the corresponding adjacent normal gastric mucosa, suggesting
that miR-146a acted as a tumor suppressor gene. Early detection of
the expression of miR-146a of gastric cancer may assist in the
early diagnosis of the disease. In the present study, correlation
analysis between the expression of miR-146a and relevant
clinicopathological factors in gastric cancer showed that a low
expression of miR-146a was significantly associated with lymph node
metastasis in the patients. The relative expression of miR-146a in
patients with advanced gastric cancer and lymph node metastasis was
lower, compared with that in patients with advanced gastric cancer
without lymph node metastasis. These results also suggested that
miR-146a may act as a tumor suppressor, which increased the degree
of malignancy and led to early metastasis of the tumor. As tumors
progressed between early stages (stage I and II) and late stages
(stage III and IV), the expression of miR-146a decreased, although
no statistical significance was found. Previous studies also showed
that the expression of miRs in patient tumor tissue had no
significant correlation with the clinical staging of advanced
gastric cancer (28,29), which was also observed in tissue
samples. Further studies are required to investigate the reasons
behind this discrepancy. In the present study, the expression of
miR-146a was correlated with patient survival rates, which was
consistent with the findings of a previous study on gastric cancer
(8). Compared with previous findings,
the present study showed that the expression of miR-146a was
associated with early diagnosis of gastric cancer, and also had
specificity in the diagnosis of gastric cancer. The present study
also found that the expression of miR-146a was associated with the
efficacy of anticancer treatment. Patients with high expression
levels of miR-146a in advanced gastric cancer tissues demonstrated
a significantly higher treatment efficacy.
According to biological prediction, miR-146a binds
to the 3′UTR of LIN52 to regulate the expression of LIN52 and
affect LIN52 function. The immunohistochemical staining showed that
the protein expression of LIN52 in advanced gastric cancer tissue
was negatively correlated with the expression of miR-146a,
suggesting that LIN52 was involved as an oncogene, compared with
miR-146a. A previous study showed that inhibition of the expression
of LIN52 in gastrointestinal stromal tumors promoted
imatinib-induced apoptosis and was involved in tumor suppression
(9). The present study found that the
inhibition of LIN52 by miR-146a resulted in improved survival rates
of the patients. Several drugs used for chemotherapy have poor
efficacy against tumor cells at the G0 phase of the cell cycle.
However, numerous tumor cells are at the G0 phase, which is also a
major reason for the resistance to drug chemotherapy (30). The activation of LIN52 directly
affects the cell cycle, allowing the majority of cells to remain at
the G0 resting phase (31) and
preventing the cells from entering the DNA synthesis phase (S
phase) (32). miR-146a inhibits the
expression of LIN52 and reduces the number of cancer cells
remaining in the G0 phase, thereby increasing the ratio of cancer
cells at the S phase and improving the efficacy of anticancer
therapy. A previous study showed that miRs affect the cell cycle of
cancer cells, which may be one of the reasons they can affect the
resistance of tumors to drugs (33).
Although the present study showed that the expression of miR-146a
was significantly correlated with the expression of LIN52 and with
chemotherapeutic sensitivity, it is unclear whether miR-146a
affects and reverses the effects of chemotherapy through LIN52 to
regulate cancer cell cycle. Further investigations are required to
verify these mechanisms.
Although miRs are promising markers in the diagnosis
and prognosis of cancer, and dozens of miRs can be used for the
diagnosis of gastric cancer (34,35), there
are several restrictions on methods and technical limitations in
the clinical detection of miRs. Currently, several detection
methods and software packages are available for miR detection in
different types of cancer. However, standardized approaches are
different, leading to inconsistent findings among studies (36). For this reason, it is necessary to
develop standardized methods of assessment and introduce
housekeeping miRs with stable expression, including miR-16 and
RUN6B. For the detection of gastric cancer, serum miR-93 is
recommended as a marker gene to identify healthy controls (37). Others have also suggested the use of
miRs with low expression levels in humans but high expression in
lower organisms, including Caenorhabditis elegans, as
internal controls (38). With
advancements in the future, microRNAs are likely to be of increased
clinical use and offer more accurate guidance in cancer
investigations.
Acknowledgements
This study was supported by the Natural Science
Foundation of China (grant no. U1504820).
References
|
1
|
Yang T, Zeng H, Chen W, Zheng R, Zhang Y,
Li Z, Qi J, Wang M, Chen T, Lou J, et al: Helicobacter pylori
infection, H19 and LINC00152 expression in serum and risk of
gastric cancer in a Chinese population. Cancer Epidemiol.
44:147–153. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Colquhoun A, Arnold M, Ferlay J, Goodman
KJ, Forman D and Soerjomataram I: Global patterns of cardia and
non-cardia gastric cancer incidence in 2012. Gut. 64:1881–1882.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sempere LF: Integrating contextual miRNA
and protein signatures for diagnostic and treatment decisions in
cancer. Expert Rev Mol Diagn. 11:813–827. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Imam JS, Buddavarapu K, LeeChang JS,
Ganapathy S, Camosy C, Chen Y and Rao MK: MicroRNA-185 suppresses
tumor growth and progression by targeting the Six1 oncogene in
human cancers. Oncogene. 29:4971–4979. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin ZW, Jiang W and Wang L: Biomarkers for
gastric cancer: Progression in early diagnosis and prognosis
(Review). Oncol Lett. 9:1502–1508. 2015.PubMed/NCBI
|
|
6
|
Yao Q, Cao Z, Tu C, Zhao Y, Liu H and
Zhang S: MicroRNA-146a acts as a metastasis suppressor in gastric
cancer by targeting WASF2. Cancer Lett. 335:219–224. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hou Z, Xie L, Yu L, Qian X and Liu B:
MicroRNA-146a is down-regulated in gastric cancer and regulates
cell proliferation and apoptosis. Med Oncol. 29:886–892. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: Clinical significance of miR-146a in gastric cancer cases.
Clin Cancer Res. 17:4277–4284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boichuk S, Parry JA, Makielski KR,
Litovchick L, Baron JL, Zewe JP, Wozniak A, Mehalek KR,
Korzeniewski N, Seneviratne DS, et al: The DREAM complex mediates
GIST cell quiescence and is a novel therapeutic target to enhance
imatinib-induced apoptosis. Cancer Res. 73:5120–5129. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grimson A, Farh KK, Johnston WK,
GarrettEngele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jensen EH and Tuttle TM: Preoperative
staging and postoperative surveillance for gastric cancer. Surg
Oncol Clin N Am. 16:329–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duan X, Cao W, Wang L, Liu S, Liu Z, Zhang
B, Yang H, Feng T, Zhang J, Zhang X, et al: Genetic variants in
TERT are associated with risk of gastric cancer in a Chinese Han
population. Oncotarget. Nov 4–2016.(Epub ahead of print).
View Article : Google Scholar
|
|
13
|
Takahashi TY, Saikawa Y and Kitagawa Y:
Gastric cancer: Current status of diagnosis and treatment. Cancers
(Basel). 5:48–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsumoto K, Ueyama H, Matsumoto K,
Akazawa Y, Komori H, Takeda T, Murakami T, Asaoka D, Hojo M, Tomita
N, et al: Clinicopathological features of alpha-fetoprotein
producing early gastric cancer with enteroblastic differentiation.
World J Gastroenterol. 22:8203–8210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He CZ, Zhang KH, Li Q, Liu XH, Hong Y and
Lv NH: Combined use of AFP, CEA, CA125 and CAl9-9 improves the
sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol.
13:872013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu WY, Xue XY, Chen ZJ, Han SL, Huang YP,
Zhang LF, Zhu GB and Shen X: Potentially predictive microRNAs of
gastric cancer with metastasis to lymph node. World J
Gastroenterol. 17:3645–3651. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu HS and Xiao HS: MicroRNAs as potential
biomarkers for gastric cancer. World J Gastroenterol.
20:12007–12017. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui L, Zhang X, Ye G, Zheng T, Song H,
Deng H, Xiao B, Xia T, Yu X, Le Y and Guo J: Gastric juice
MicroRNAs as potential biomarkers for the screening of gastric
cancer. Cancer. 119:1618–1626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu JL, Gao W, Kang QM, Zhang XJ and Yang
SG: Prognostic value of survivin in patients with gastric cancer: A
systematic review with meta-analysis. PLoS One. 8:e719302013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cerne JZ, Gersak K and Novakovic S: The
influence of the genetic variant within miRNA-binding site in
estrogen receptor alpha gene on the risk of breast cancer in
postmenopausal women on hormone replacement therapy. Cancer
Biomark. 8:123–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim BH, Hong SW, Kim A, Choi SH and Yoon
SO: Prognostic implications for high expression of oncogenic
microRNAs in advanced gastric carcinoma. J Surg Oncol. 107:505–510.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Song YX and Wang ZN: Non-coding
RNAs in gastric cancer. Gene. 560:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zandberga E, Kozirovskis V, Ābols A,
Andrējeva D, Purkalne G and Linē A: Cell-free microRNAs as
diagnostic, prognostic, and predictive biomarkers for lung cancer.
Genes Chromosomes Cancer. 52:356–369. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Verma M, Lam TK, Hebert E and Divi RL:
Extracellular vesicles: Potential applications in cancer diagnosis
prognosis, and epidemiology. BMC Clin Pathol. 15:62015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Migliore C and Giordano S: Resistance to
targeted therapies: A role for microRNAs? Trends Mol Med.
19:633–642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rosenfeld N, Aharonov R, Meiri E,
Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S,
Levy A, et al: MicroRNAs accurately identify cancer tissue origin.
Nat Biotechnol. 26:462–429. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY
and Zhu ZG: MiRNA-199a-3p: A potential circulating diagnostic
biomarker for early gastric cancer. J Surg Oncol. 108:89–92. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cai H, Yuan Y, Hao YF, Guo TK, Wei X and
Zhang YM: Plasma microRNAs serve as novel potential biomarkers for
early detection of gastric cancer. Med Oncol. 30:4522013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shah MA and Schwartz GK: Cell
cycle-mediated drug resistance: An emerging concept in cancer
therapy. Clin Cancer Res. 7:2168–2181. 2001.PubMed/NCBI
|
|
31
|
Litovchick L, Florens LA, Swanson SK,
Washburn MP and DeCaprio JA: DYRK1A protein kinase promotes
quiescence and senescence through DREAM complex assembly. Genes
Dev. 25:801–813. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sadasivam SS, Duan S and DeCaprio JA: The
MuvB complex sequentially recruits B-Myb and FoxM1 to promote
mitotic gene expression. Genes Dev. 26:474–489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xie L, Jing R, Qi J, Lin Z and Ju S: Drug
resistance-related microRNAs in hematological malignancies:
Translating basic evidence into therapeutic strategies. Blood Rev.
29:33–44. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rotkrua P, Shimada S, Mogushi K, Akiyama
Y, Tanaka H and Yuasa Y: Circulating microRNAs as biomarkers for
early detection of diffuse-type gastric cancer using a mouse model.
Br J Cancer. 108:932–940. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tsujiura M, Komatsu S, Ichikawa D,
Shiozaki A, Konishi H, Takeshita H, Moriumura R, Nagata H,
Kawaguchi T, Hirajima S, et al: Circulating miR-18a in plasma
contributes to cancer detection and monitoring in patients with
gastric cancer. Gastric Cancer. 18:271–279. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tiberio P, Callari M, Angeloni V, Daidone
MG and Appierto V: Challenges in using circulating miRNAs as cancer
biomarkers. Biomed Res Int. 2015:7314792015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song J, Bai Z, Han W, Zhang J, Meng H, Bi
J, Ma X, Han S and Zhang Z: Identification of suitable reference
genes for qPCR analysis of serum microRNA in gastric cancer
patients. Dig Dis Sci. 57:897–904. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods.
50:298–301. 2010. View Article : Google Scholar : PubMed/NCBI
|