Introduction
Helicobacter pylori infection of the gastric
mucosa results in gastritis in the majority of infected individuals
(1). This infection is a major cause
of peptic ulcer disease (PUD) and an important causative factor for
gastric cancer (GC) (1). GC is the
second most common cause of cancer-associated mortalities and is
one of the most prevalent malignancies worldwide (2). H. pylori-induced progression
towards PUD or GC is likely to depend upon a combination of several
factors, including the bacterial genotype, host susceptibility,
immune response and environmental factors.
Cytotoxin-associated gene A (CagA) is a 120-145-kDa
protein produced by H. pylori and is the product of the
cagA gene (3,4). The cagA gene is not present in
all H. pylori strains and the majority of studies report an
increased risk of PUD and GC with CagA-positive H. pylori
strains (5,6). In patients infected with CagA-positive
H. pylori strains, the CagA cytotoxin is injected into the
epithelial cells of the stomach. The C-terminal region of CagA
varies among H. pylori strains and is the target region for
tyrosine phosphorylation by host cell kinases (7). Four tyrosine phosphorylation motifs
(EPIYA) are located within this region and have been characterized
with respect to the amino acid sequences flanking these sites
(7,8).
These four sites, termed EPIYA-A, -B, -C and -D, vary in number and
organization in different H. pylori strains (7,8). Based on
the arrangement of the EPIYA motifs, there are two types of CagA
designated Western CagA and East Asian CagA (8). The Western type CagA has EPIYA-A and
EPIYA-B sites followed by 1 to 3 EPIYA-C sites, while the East
Asian CagA has EPIYA-A and EPIYA-B sites, but lacks EPIYA-C
(8). The third domain of the East
Asian CagA is EPIYA-D (8). Regarding
clinical significance, the East Asian CagA is considered to be more
virulent than the Western type. For the Western CagA, an increased
number of EPIYA-C motifs is associated with greater pathogenicity
(8).
Within the variable region of the cagA gene
is another motif called the CagA multimerization (CM) region
(9). This is a 16-amino acid
sequence, which is responsible for dimerization of CagA (9). Similar to the EPIYA patterns, Western
and Eastern CM sequences have been identified (10). However, unlike CagA EPIYA motifs, the
frequency and arrangement of Western and Eastern CM motifs and
their association with clinical outcomes has not been well studied.
In Western CagA proteins, a CM motif is often located prior to and
following the EPIYA-C motif. A previous study examined H.
pylori infection in a minority patient population in New York
City and observed that this population had a higher infection rate
than the general population (11).
Furthermore, this population is reportedly infected with more
virulent CagA-positive H. pylori strains that are associated
with an increased risk of precancerous changes in the gastric
mucosa (12).
A subsequent study examined cagA genes from
H. pylori strains infecting the same patient population from
New York City (13). It was reported
that all CagA proteins were of the Western type with respect to
EPIYA motifs and the majority of the EPIYA motifs were arranged in
an ABC pattern (13) However, a
greater degree of heterogeneity in the samples with respect to the
CM motifs surrounding the EPIYA-C phosphorylation motif was
observed (13). Notably, analysis of
the CM types and arrangements in these samples suggests that the
presence of one or two Western CM motifs (designated WW or W-) in
the absence of an Eastern CM motif is associated with more severe
gastric disorders like PUD and GC (13). Consistent with these results, H.
pylori strains expressing Western CagA proteins with the
Western/Western CM motif pattern have been associated with a
greater mean length of cell elongation and a greater affinity for
SHP-2 when compared with CagA proteins with the Western/Eastern
pattern (10). This suggests that in
Western CagA proteins, which contain the ABC EPIYA pattern, an
Eastern CM motif may decrease CagA multimerization and virulence.
Therefore, in H. pylori-infected patients that are
cagA-positive, the pattern and types of CM motifs may be
clinically significant.
Although the CM regions may affect the magnitude of
the pathophysiological activities of CagA, the effect of particular
CM sequences and arrangements on cell function has not yet been
studied. The present study focused specifically on the effects of
CagA proteins with Western/Western and Western/Eastern CM patterns
on gastric cell function using human gastric adenocarcinoma (AGS)
cells, a human gastric tumor cell line. The results suggest that
different CM motif types and arrangements may affect CagA
virulence.
Materials and methods
Cloning and mutagenesis of the CagA
C-terminal region
The C-terminal region of Western CagA proteins
usually contain at least three EPIYA motifs (EPIYA-A, EPIYA-B and
EPIYA-C), in addition to two CM motifs flanking the EPIYA-C motif
(7–9).
The CagA C-terminal region obtained from a H.
pylori-infected subject from a previous study (13) was cloned and contained the terminal
386 amino acids of the CagA protein (based on CagA sequence
CBV36576). This CagA C-terminal fragment was amplified by
polymerase chain reaction and cloned into the pAcGFP1-C3 vector
(Clontech Laboratories, Inc., Mountainview, CA, USA) downstream to
and in frame with green fluorescent protein (GFP). The primers used
were as follows: CagA-DF, GGA
TTGAGCTCATGGGAACAGGCGATTTCAGTGGGGTAG and CagA-DR, CCG
AGGTACCTTAAGATTTTTGGAAACCACCTTTTGTA (14). The CagA-DF and CagA-DR primers
contained SacI and KpnI restriction sites (italicized
above), respectively.
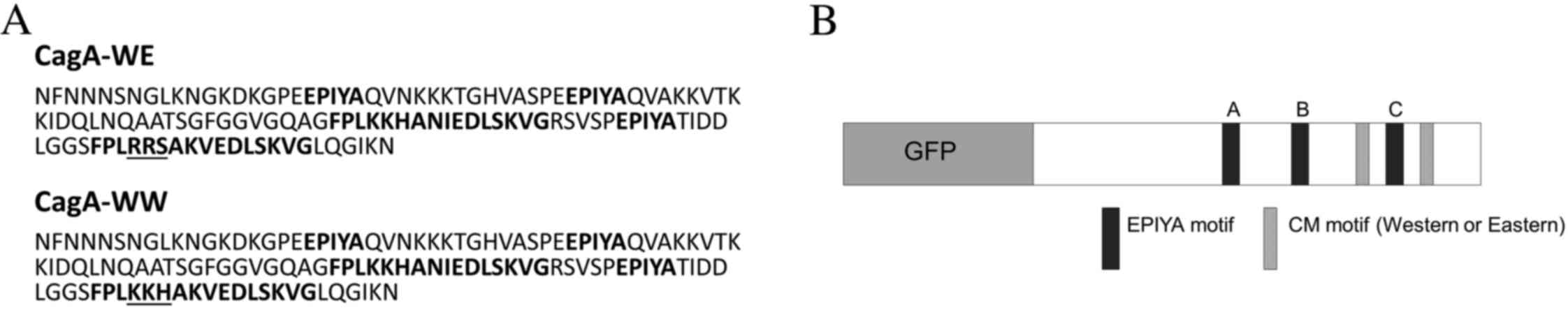
The portion of the cloned peptide containing the
EPIYA and CM motifs is presented in Fig.
1A and contains three EPIYA phosphorylation motifs (A, B and
C), in addition to Western and Eastern CM motifs flanking the
EPIYA-C region. The amino acid sequence of the underlined portion
of the Eastern CM motif (Fig. 1A) was
mutated from RRS to KKH, and subsequently resembled a Western CM
motif based on CM motifs observed in strains from Eastern or
Western countries (10). The sequence
integrity of each construct was verified by DNA sequencing. The
result was two GFP-CagA constructs that were identical except for
the CM motifs flanking the EPIYA-C region (Fig. 1B). One construct, GFP-CagA-WE,
contained a Western/Eastern CM motif arrangement, while the other,
GFP-CagA-WW, contained a Western/Western arrangement.
Cell culture and transfection
AGS cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and cultured in F12K media
with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA), 1X penstrep and 2 mM glutamine. Cells were grown
on T-25 flasks, 6-well plates or Nunc chamber slides and
transfected using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to manufacturer's
protocol. The DNA to Lipofectamine ratio was 1:2.5 and transfection
efficiency was >75%. Cell viability was measured using the
Trypan blue (TB) exclusion assay. Briefly, equal amounts of cell
suspension were mixed with 0.4% TB in PBS. The cells were examined
under a microscope and the percentage of TB-positive cells was
determined.
Immunoblotting of subcellular
fractions
AGS cells were transfected with plasmids containing
the GFP-CagA constructs, GFP without CagA or no plasmid. After
18–20 h, the cells were washed with PBS and harvested by scraping
into ice-cold PBS and centrifuged at 1,500 × g for 5 min at
room temperature. In certain experiments, detached cells in the
media were collected by centrifugation at 1,500 × g for 5
min at room temperature and washed with PBS. Cell pellets were
resuspended in lysing solution [20 mM Tris-HCL (pH 7.2), 150 mM
NaCl, 1% TX-100, 1 mM NaF, 2 mM Na3VO4 and 1X
Halt protease inhibitor cocktail (Thermo Fisher Scientific, Inc.)].
Protein assays were performed using the Coomassie Protein assay
reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, and immunoblotting was performed as
previously described (15). Primary
antibodies included anti-GFP (#PA1980A; Thermo Fisher Scientific,
Inc.), anti-Caspase-3 (#9662; Cell Signaling Technology, Inc.,
Danvers, MA, USA) and Ki-67 (#RB9043; Thermo Fisher Scientific,
Inc.) and were diluted at 1:1,000.
Immunocytochemistry
Cells were grown on Nunc chamber slides and
transfected as aforementioned. Following 18 h, the cells were fixed
with 4% paraformaldehyde for 10 min and washed in PBS. GFP-CagA
proteins were visualized by immunofluorescence. For Ki-67 staining,
the cells were incubated with PBS containing 3% bovine serum
albumin (BSA), 1% goat serum (Thermo Fisher Scientific, Inc.) and
0.2% Triton X-100 (PBS-BSA/GS) for 1 h followed by incubation with
anti-Ki-67 (1:1,000) overnight at 4°C. The slides were incubated
with Alexa Fluor-568-conjugated goat anti-rabbit immunoglobulin G
secondary antibodies, washed and mounted with DAPI containing
mounting media. Fluorescent signals were visualized using an SP2
confocal microscope (Leica Microsystems, Inc., Buffalo Grove, IL,
USA) or an AX10 microscope (Zeiss AG, Oberkochen, Germany). Images
were obtained, merged and assembled using Adobe Photoshop software
v7.0 (Adobe Systems, Inc., San Jose, CA, USA). To measure apoptosis
in transfected cells, the In Situ Cell Death Detection kit (Roche,
Indianapolis, IN, USA) was used according to the manufacturer's
protocol.
Cell elongation assays
CagA has been reported to induce cell elongation or
the ‘hummingbird phenotype’ in epithelial cells (16,17).
Hummingbird cells are defined as having needle-like cellular
protrusions >5 µm in length. To determine whether or not the CM
motif type and arrangement affected AGS cell morphology, the
percent of transfected cells exhibiting this phenotype was
determined. At least 200 cells were counted per transfectant in
each experiment. The average length of the cell protrusions of at
least 50 cells per transfectant was measured using Spot Advanced
software v4.6 (SPOT Imaging Solutions; Diagnostic Instruments,
Inc., Sterling Heights, MI, USA) on images of captured with an AX10
microscope (Zeiss AG).
Statistical analysis
Data presented in the figures are representative of
at least three different experiments. The data are presented as the
mean ± standard error of the mean. Comparisons between groups were
evaluated by Student's t-test, and P<0.05 were considered to
indicate a statistically significant difference.
Results and Discussion
CagA constructs and expression in AGS
cells
In the present study, the effects of two CagA
C-terminal peptides with two different CM patterns on epithelial
function and morphology were examined. The only difference between
the two CagA constructs used in the current study was the CM
pattern. These constructs were cloned downstream and in frame with
GFP as described in the Materials and methods section and are
presented in Fig. 1A and B. The CagA
C-terminal fragment containing two Western type CM regions is
referred to as the CagA-WW, while the fragment containing Western
and Eastern CM motifs is referred to as the CagA-WE. The vector
containing GFP without a CagA fragment was used as a control.
Previous studies have demonstrated that the C-terminal CagA
fragment exhibits CagA activity, including the induction of cell
elongation and interleukin-8 secretion in AGS cells (14,18). This
fragment, which contains the terminal third of the CagA protein,
was observed to induce greater cell elongation than the complete
CagA protein (18).
AGS cells were transfected with plasmids coding for
GFP, GFP-CagA-WE, GFP-CagA-WW or no plasmid. After 18 h, lysates
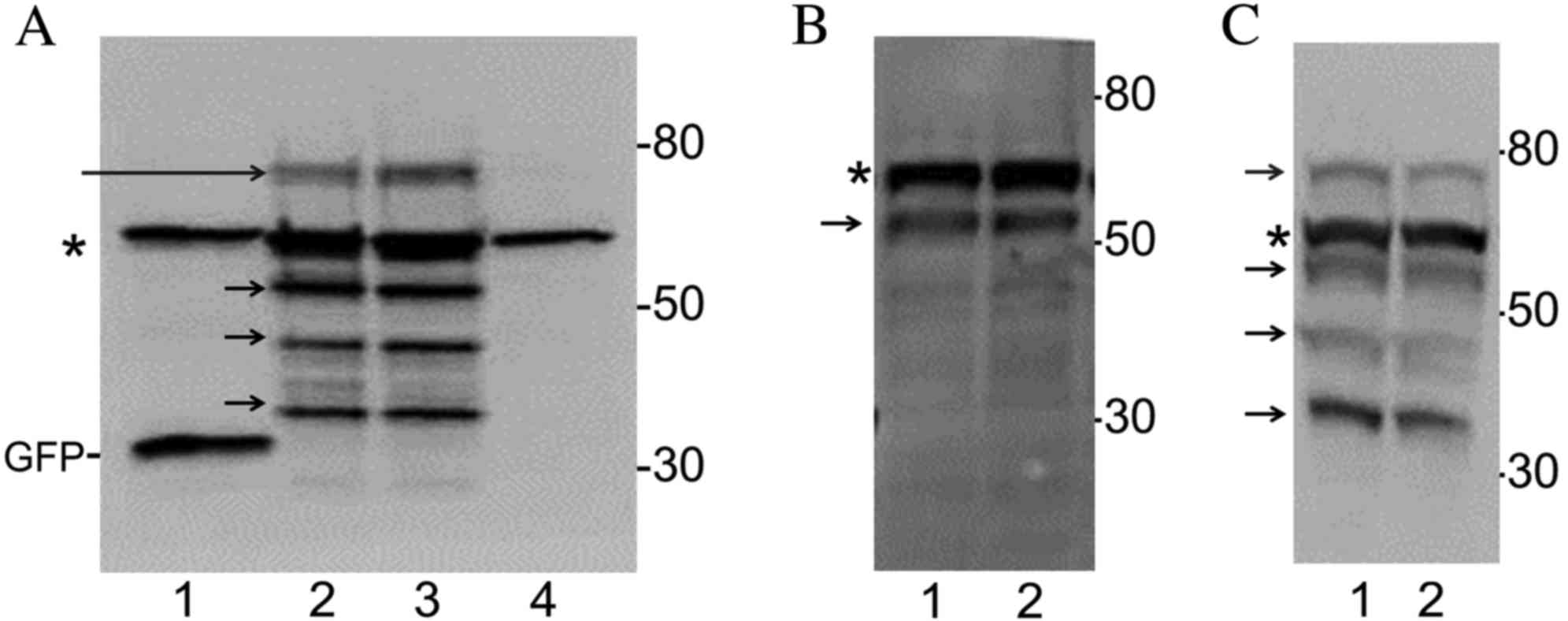
were prepared and immunoblotted with an anti-GFP antibody. As
presented in Fig. 2A (lane 1), a
27-kD band was observed in lysates from cells transfected with the
GFP plasmid, which corresponds with the molecular weight (MW) of
GFP. In cells transfected with no plasmid, no new bands were
observed (Fig. 2A, lane 4). By
contrast, an ~70-kD band was observed in lysates prepared from
cells expressing either GFP-CagA-WW or GFP-CagA-WE (Fig. 2A, lanes 2 and 3, respectively). The
70-kD band was observed just above a non-specific band present in
all four samples. Three additional low MW bands were observed in
lysates prepared from cells expressing either GFP-CagA-WW or
GFP-CagA-WE at ~55 kDa, 40 kDa and 35 kDa in lanes 2 and 3
(Fig. 2A). These may represent
cleaved fragments of the GFP-CagA peptide. Fragmentation of
GFP-CagA was observed to varying degrees in cell lysates from
different transfection experiments (Fig.
2B and C). The level of expression of GFP-CagA-WW and
GFP-CagA-WE was similar in cells transfected with these
plasmids.
Cleavage of CagA following infection of epithelial
cells with H. pylori has been observed previously and it was
suggested that CagA cleavage may be involved with host signal
transduction and virulence (19,20).
Notably, CagA cleavage is not observed in bacterial cells, and
occurs only following translocation into the host cell (20). It is unlikely that cleavage of the
GFP-CagA protein occurred as a result of cell manipulation as
lysates were prepared in buffers containing protease and
phosphatase inhibitors and cold reagents, and, as aforementioned,
the GFP remained intact. Whether or not CagA requires fragmentation
to function remains to be determined.
In the current study, attempts were made to use a
commercially prepared CagA-specific antibody to immunoblot lysates
prepared from GFP-CagA expressing cells. However, this antibody did
not recognize the GFP-CagA proteins. This was most likely due to
the fact that the C-terminal is the most variable region in the
CagA protein and may not be recognized by commercially prepared
antibodies. Lack of antigenicity in cells expressing the CagA
C-terminal fragment has been observed previously (14). Nevertheless, the sequences of each
GFP-CagA construct were analyzed and it was determined that the
CagA sequences were in frame with the GFP sequence in the plasmid.
Furthermore, the size of the high MW band recognized by the GFP
antibody was consistent with the calculated MW of the GFP-CagA
constructs, which was ~69 kD.
CagA expression affects AGS cell
adhesion
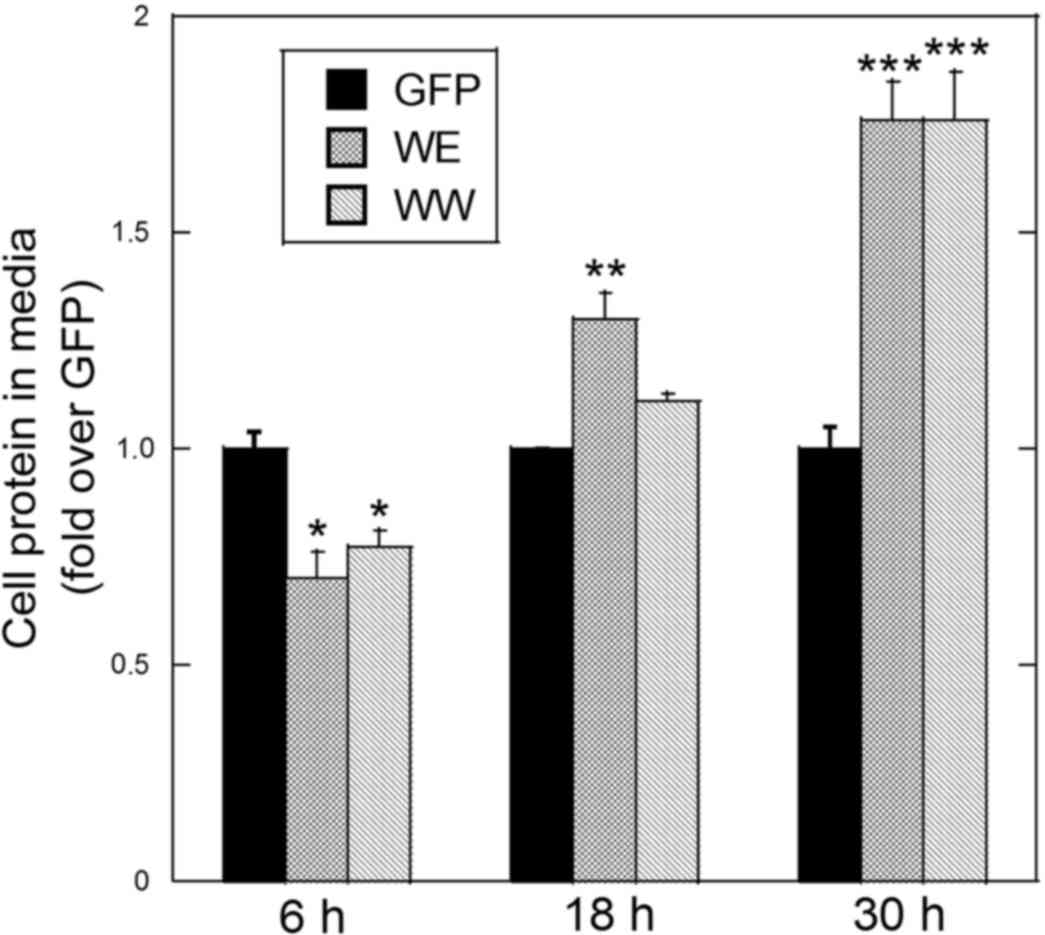
In the present study, an apparent increase in
detached cells in AGS cells expressing either GFP-CagA-WE or
GFP-CagA-WW for 18 h or longer was observed. To quantitate the
effect of CagA on cell adhesion, the cells were transfected and
grown for 6, 18 or 30 h. The protein levels of adherent and
detached cells were determined and expressed as percent cellular
protein in media. The detached cells were washed in PBS to rid the
sample of secreted proteins or proteins derived from the FBS added
to the media. The values were normalized to the protein level in
detached cells transfected with the GFP plasmid. At 6 h
post-transfection, a lower level of cellular protein in the media
was observed with cells expressing either GFP-CagA-WE or
GFP-CagA-WW compared with cells expressing GFP alone (Fig. 3). This suggests that CagA-transfected
cells were more adherent at this early time point. At 18 h
post-transfection, a higher level of protein in the media was
observed with cells expressing either GFP-CagA-WE or GFP-CagA-WW
compared with cells expressing GFP alone (Fig. 3). Furthermore, at this time point the
amount of cellular protein in the media was higher in GFP-CagA-WE
expressing cells compared with GFP-CagA-WW expressing cells
(Fig. 3). At 36 h post-transfection,
protein levels in the media were almost two-fold higher in cells
expressing either GFP-CagA-WE or GFP-CagA-WW compared with cells
expressing GFP alone (Fig. 3). The
viability of the detached cells was >85% in all cases indicating
that these are not cells that died and detached from the culture
surface. These results suggest that different CM motif patterns
affect the rate at which the cells attach to the surface and that
each form of CagA inhibits cell attachment at time points beyond 18
h.
Other researchers have also observed decreased cell
adhesion in cells transfected with CagA or infected with
CagA+ H. pylori strains (21). This is not remarkable since it has
been demonstrated that H. pylori infection with
CagA+ strains perturbs various cell junction types,
including tight junctions (22,23), focal
adhesion complexes (24,25) and gap junctions (26). The results of the current study
indicate that overexpression of CagA eventually inhibits cell
attachment and that the effect of CagA on cell attachment is
time-dependent. Additionally, CagA-WE appears to inhibit cell
attachment sooner (18 h post-transfection). Further studies are
required to determine how the different CagA CM regions affect
specific cell junction types and how this relates to gastric
pathology.
Effect of GFP-CagA on cell
elongation
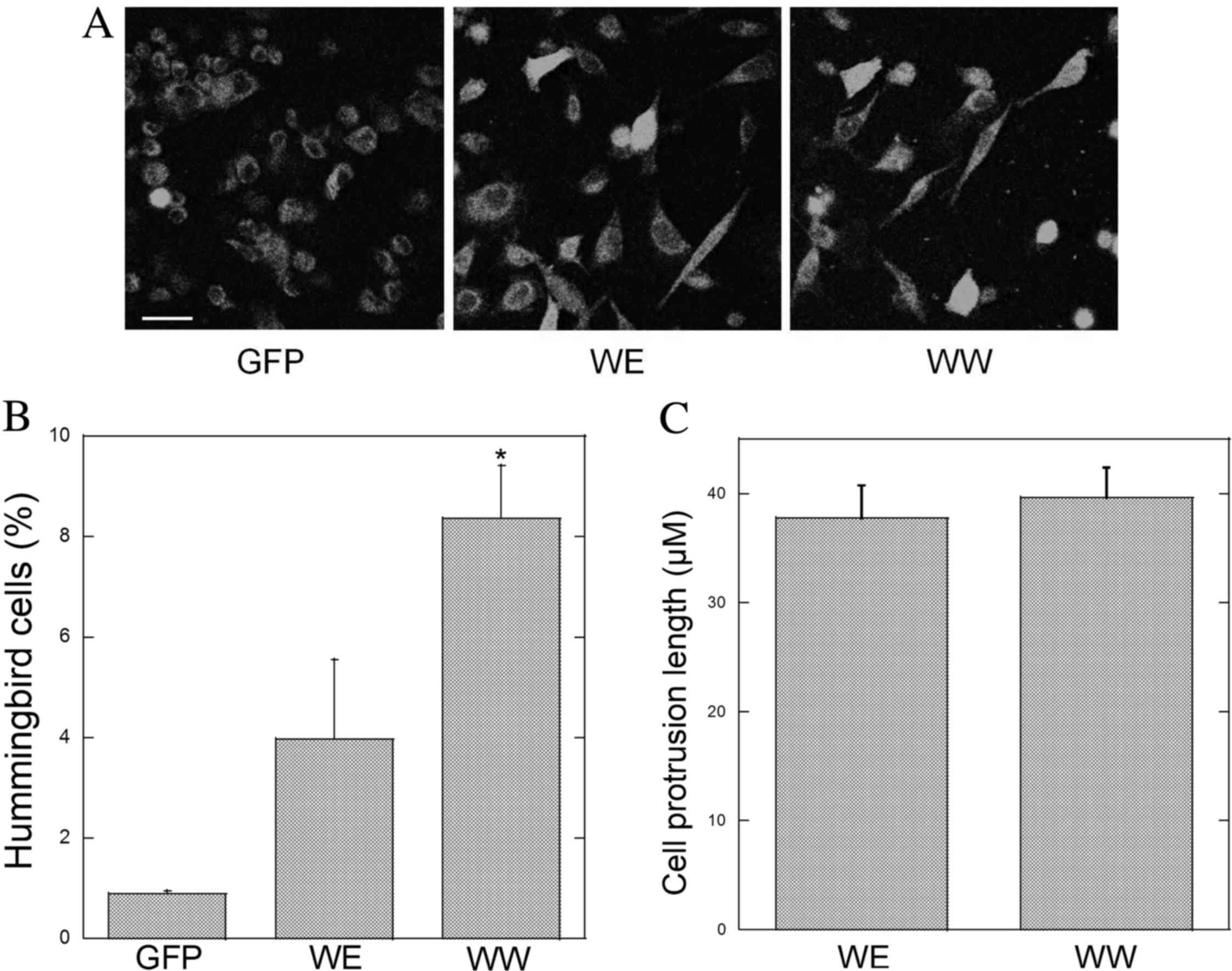
Cell elongation or the ‘hummingbird’ phenotype is a
well-accepted in vitro measure of CagA virulence and is
associated with the ability of CagA to interact with SHP-2
(17,19). To examine the effects of the different
CM motifs on cell elongation, AGS cells were transfected with GFP,
GFP-CagA-WE or GFP-CagA-WW containing plasmids. Following 18 h, the
cells were fixed and mounted. When examined at higher power by
confocal microscopy, the appearance of needle-like cytoplasmic
projections was more apparent in GFP-CagA expressing cells
(Fig. 4A). In GFP-expressing cells,
the staining pattern was cytoplasmic. By contrast, the staining
appeared to be cytoplasmic and nuclear in cells expressing
GFP-CagA-WE or GFP-CagA-WW. Note that the staining intensity of the
green fluorescence appeared to be similar in cells transfected with
the GFP, CagA-WE and CagA-WW plasmids.
The percentage of elongated cells in transfected AGS
cells was determined. A higher percentage of elongated cells was
observed with either GFP-CagA construct compared with cells
expressing GFP alone (Fig. 4B).
Notably, the highest percentage of elongated cells was observed in
cells expressing GFP-CagA-WW (Fig.
4B). However, the average length of the cell protrusions was
not different in GFP-CagA-WE or GFP-CagA-WW expressing cells
(Fig. 4C).
Taken together, these results suggest that the WW CM
motif pattern is more virulent than the WE pattern, which is
consistent with the observations of a previous study (10). However, in this previous study,
various CagA proteins were used that differed in EPIYA motifs and
other portions of the CagA sequence, whereas the results of the
present study may be attributed solely to variations in the CM
region as this was the only variable region in the CagA constructs.
Therefore, to the best of our knowledge, the current study has
demonstrated for the first time that the CM pattern affects the
ability of CagA to induce cell elongation.
Effect of GFP-CagA on cell
proliferation and apoptosis
The effect of CagA protein expression on cell
proliferation in AGS cells was measured using Ki-67 antibodies as
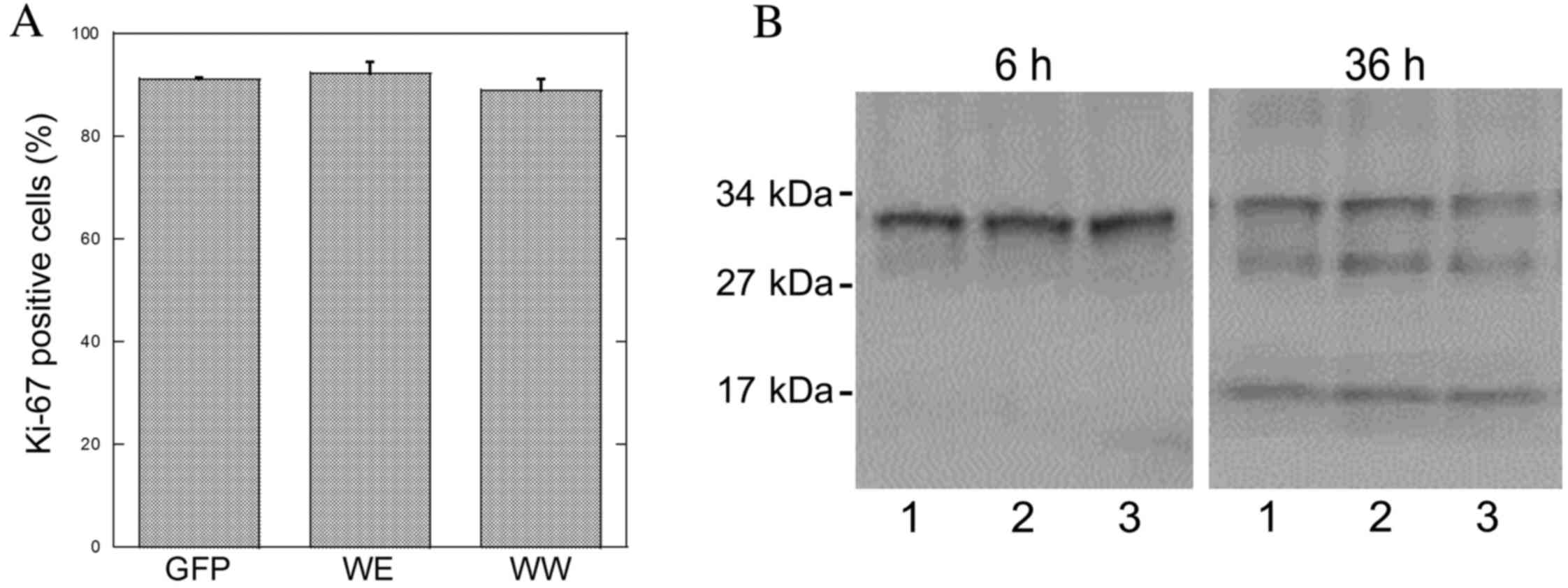
an indicator of cell proliferation. The level of labeling with
Ki-67 was ~90% and did not vary in GFP, GFP-CagA-WE or GFP-CagA-WW
expressing AGS cells (Fig. 5A). Few
apoptotic cells were detected using the TUNEL assay and due to the
extremely low level of apoptotic activity, accurate apoptotic
measurements could not be obtained (data not shown). Nevertheless,
the level of apoptosis as measured by the TUNEL assay did not
appear different in GFP-CagA-WE or GFP-CagA-WW expressing cells.
Apoptotic activity was also examined in transfected AGS cells by
immunoblotting with the caspase-3 antibody. The cleaved or active
form of caspase-3 (17 kDa) was not present in transfected cells at
6 or 18 h post-transfection (Fig.
5B). By contrast, the caspase-3 fragment was visible 36 h
post-transfection in GFP, GFP-CagA-WE or GFP-CagA-WW expressing
cells (Fig. 5B). However, the levels
of the cleaved caspase-3 fragment were not different in cells
transfected with the GFP- or CagA-containing plasmids indicating
that expression of the CagA peptide does not affect apoptotic rates
under these conditions.
Depending on the type of assay, H. pylori
infection has been associated with increased or decreased
proliferation (27–29). The CagA constructs utilized in the
present study did not affect the rate of proliferation or apoptosis
in AGS cells regardless of the CM pattern. It is possible that
these effects require long-term expression of the CagA protein,
which would be observed in chronically infected tissue, or that
other H. pylori proteins are required to affect
proliferation and/or apoptosis.
In conclusion, to the best of our knowledge, the
current study demonstrated for the first time that heterogeneity in
the CM motif patterns of the CagA virulence factor affects cell
adhesion and morphology. Further studies are required to elucidate
the mechanisms underlying these effects. Furthermore, as there is a
significant degree of heterogeneity in CM motifs from different
H. pylori strains, closer investigation into how these
differences affect CagA virulence and clinical outcomes is
warranted.
Acknowledgements
The present study was supported in part by grants
from PSC-CUNY, New York, NY, USA (no. 64464-0042), The Hunter
College Presidential Fund for Faculty Advancement, New York, NY,
USA (no. 94806-6013) and The Shuster Faculty Fellowship Fund, New
York, NY, USA (no. 78742).
References
|
1
|
Qadri Q, Rasool R, Gulzar GM, Naqash S and
Shah ZA: H. pylori infection, inflammation and gastric cancer. J
Gastrointest Cancer. 45:126–132. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Covacci A, Censini S, Bugnoli M, Petracca
R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z and Figura N:
Molecular characterization of the 128-kDa immunodominant antigen of
Helicobacter pylori associated with cytotoxicity and duodenal
ulcer. Proc Natl Acad Sci USA. 90:5791–5795. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tummuru MK, Cover TL and Blaser MJ:
Cloning and expression of a high-molecular-mass major antigen of
Helicobacter pylori: Evidence of linkage to cytotoxin production.
Infect Immun. 61:1799–1809. 1993.PubMed/NCBI
|
|
5
|
Brenner H, Arndt V, Stegmaier C, Ziegler H
and Rothenbacher D: Is Helicobacter pylori infection a necessary
condition for noncardia gastric cancer? Am J Epidemiol.
159:252–258. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hatakeyama M and Higashi H: Helicobacter
pylori CagA: A new paradigm for bacterial carcinogenesis. Cancer
Sci. 96:835–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stein M, Bagnoli F, Halenbeck R, Rappuoli
R, Fantl WJ and Covacci A: c-Src/Lyn kinases activate Helicobacter
pylori CagA through tyrosine phosphorylation of the EPIYA motifs.
Mol Microbiol. 43:971–980. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Higashi H, Tsutsumi R, Fujita A, Yamazaki
S, Asaka M, Azuma T and Hatakeyama M: Biological activity of the
Helicobacter pylori virulence factor CagA is determined by
variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci
USA. 99:14428–14433. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren S, Higashi H, Lu H, Azuma T and
Hatakeyama M: Structural basis and functional consequence of
Helicobacter pylori CagA multimerization in cells. J Biol Chem.
281:32344–32352. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sicinschi LA, Correa P, Peek RM, Camargo
MC, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Delgado A,
Mera R, et al: CagA C-terminal variations in Helicobacter pylori
strains from Colombian patients with gastric precancerous lesions.
Clin Microbiol Infect. 16:369–378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Straus EW, Patel H, Chang J, Gupta RM,
Sottile V, Scirica J, Tarabay G, Iyer S, Samuel S and Raffaniello
RD: H. pylori infection and genotyping in patients undergoing upper
endoscopy at inner city hospitals. Dig Dis Sci. 47:1575–1581. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schneller J, Gupta R, Mustafa J,
Villanueva R, Straus EW and Raffaniello RD: Helicobacter pylori
infection is associated with a high incidence of intestinal
metaplasia in the gastric mucosa of patients at inner-city
hospitals in New York. Dig Dis Sci. 51:1801–1809. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ogorodnik E and Raffaniello RD: Analysis
of the 3′-variable region of the cagA gene from Helicobacter pylori
strains infecting patients at New York City hospitals. Microb
Pathog. 56:29–34. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim SY, Lee YC, Kim HK and Blaser MJ:
Helicobacter pylori CagA transfection of gastric epithelial cells
induces interleukin-8. Cell Microbiol. 8:97–106. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Limi S, Ojakian G and Raffaniello R: Rab3D
regulates amylase levels, not agonist-induced amylase release, in
AR42 J cells. Cell Mol Biol Lett. 17:258–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Higashi H, Nakaya A, Tsutsumi R, Yokoyama
K, Fujii Y, Ishikawa S, Higuchi M, Takahashi A, Kurashima Y,
Teishikata Y, et al: Helicobacter pylori CagA induces
Ras-independent morphogenetic response through SHP-2 recruitment
and activation. J Biol Chem. 279:17205–17216. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Segal ED, Cha J, Lo J, Falkow S and
Tompkins LS: Altered states: Involvement of phosphorylated CagA in
the induction of host cellular growth changes by Helicobacter
pylori. Proc Natl Acad Sci USA. 96:14559–14564. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pelz C, Steininger S, Weiss C, Coscia F
and Vogelmann R: A novel inhibitory domain of Helicobacter pylori
protein CagA reduces CagA effects on host cell biology. J Biol
Chem. 286:8999–9008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Backert S, Muller EC, Jungblut PR and
Meyer TF: Tyrosine phosphorylation patterns and size modification
of the Helicobacter pylori CagA protein after translocation into
gastric epithelial cells. Proteomics. 1:608–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Moese S, Selbach M, Zimny-Arndt U,
Jungblut PR, Meyer TF and Backert S: Identification of a
tyrosine-phosphorylated 35 kDa carboxy-terminal fragment (p35CagA)
of the Helicobacter pylori CagA protein in phagocytic cells:
Processing or breakage? Proteomics. 1:618–629. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alfizah H and Ramelah M: Variant of
Helicobacter pylori CagA proteins induce different magnitude of
morphological changes in gastric epithelial cells. Malays J Pathol.
34:29–34. 2012.PubMed/NCBI
|
|
22
|
Amieva MR, Vogelmann R, Covacci A,
Tompkins LS, Nelson WJ and Falkow S: Disruption of the epithelial
apical-junctional complex by Helicobacter pylori CagA. Science.
300:1430–1434. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song X, Chen HX, Wang XY, Deng XY, Xi YX,
He Q, Peng TL, Chen J, Chen W, Wong BC and Chen MH: H.
pylori-encoded CagA disrupts tight junctions and induces
invasiveness of AGS gastric carcinoma cells via Cdx2-dependent
targeting of Claudin-2. Cell Immunol. 286:22–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moese S, Selbach M, Brinkmann V, Karlas A,
Haimovich B, Backert S and Meyer TF: The Helicobacter pylori CagA
protein disrupts matrix adhesion of gastric epithelial cells by
dephosphorylation of vinculin. Cell Microbiol. 9:1148–1161. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schneider S, Weydig C and Wessler S:
Targeting focal adhesions: Helicobacter pylori-host communication
in cell migration. Cell Commun Signal. 6:22008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tao R, Hu MF, Lou JT and Lei YL: Effects
of H. pylori infection on gap-junctional intercellular
communication and proliferation of gastric epithelial cells in
vitro. World J Gastroenterol. 13:5497–5500. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peek RM Jr, Moss SF, Tham KT, Pérez-Pérez
GI, Wang S, Miller GG, Atherton JC, Holt PR and Blaser MJ:
Helicobacter pylori cagA+ strains and dissociation of gastric
epithelial cell proliferation from apoptosis. J Natl Cancer Inst.
89:863–868. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ricci V, Ciacci C, Zarrilli R, Sommi P,
Tummuru MK, Del Vecchio Blanco C, Bruni CB, Cover TL, Blaser MJ and
Romano M: Effect of Helicobacter pylori on gastric epithelial cell
migration and proliferation in vitro: Role of VacA and CagA. Infect
Immun. 64:2829–2833. 1996.PubMed/NCBI
|
|
29
|
Rudnicka W, Covacci A, Wadstrom T and
Chmiela M: A recombinant fragment of Helicobacter pylori CagA
affects proliferation of human cells. J Physiol Pharmacol.
49:111–119. 1998.PubMed/NCBI
|