Introduction
Lung cancer, mainly non-small cell lung cancer
(NSCLC), is the leading cause of cancer-associated mortality
globally. More than 133,000 new diagnoses and over 77,000
associated mortalities are expected to occur in 2015 in Japan
(1). In spite of progress in NSCLC
therapy, the prognosis for patients with NSCLC remains poor
(2). Therefore, clarification of the
biomarkers most likely to predict the recurrence of NSCLC after
curative surgery is required to improve the outcome of NSCLC
patients.
MicroRNAs (miRNAs/miRs) are one of the most
compelling molecular markers in the diagnosis and prognosis of
tumors (3,4). miRNAs are short (20–24 nt) non-coding
RNAs that take part in the post-transcriptional regulation of gene
expression in multicellular organisms by impacting mRNA stability
and translation. miRNAs target protein-coding mRNAs at the
post-transcriptional level by directly cleaving mRNA or by
inhibiting protein synthesis. miRNA dysregulation has been
demonstrated to be associated with the initiation and progression
of cancer, indicating that miRNAs could be used as molecular
biomarkers for cancer diagnostics and for predicting the likely
prognosis (5,6).
Recent evidence has indicated that such miRNAs have
been identified in the plasma and serum, and are important as
non-invasive biomarkers for cancer patients (7). In NSCLC, certain studies have shown the
prognostic value of specific miRNAs in the plasma and serum
(8–11). Furthermore, a focus has been placed on
miRNAs that have also been identified in the exosome of plasma and
serum in an unexpectedly stable form, which is protected from
endogenous RNase activity (12–14).
Exosomes are small membrane vesicles (30–100 nm) whose derivation
lies in the luminal membranes of multivesicular bodies and which
are released by fusion with the cell membrane (15). It is known that exosomes transfer not
only membrane components, but also nucleic acids such as miRNAs to
other cells (16). Therefore, the
exosomal transport of miRNAs has received attention in cancer
research as a carrier of genetic information (17,18). The
early diagnosis of cancer is extremely important for improving the
prognosis. However, the existing biomarkers, such as
cancer-associated antigens, and proteomics analysis are
unsatisfactory for the early stages of NSCLC. By contrast, recent
studies have demonstrated that specific expression profiles of
circulating exosomal miRNAs could be promising biomarkers for the
detection of early-stage cancer and for predicting prognosis in
various types of cancers (19–21).
However, previous studies in NSCLC have not examined exosomal
miRNAs, which could be expected to include the intact miRNA.
The present study clarified the potential of plasma
exosomal miRNA as a biomarker for recurrence in NSCLC patients who
have undergone curative surgery.
Patients and methods
Study design
In this study, 201 NSCLC patients were included.
First, the recurrence-specific exosomal miRNAs of 6 NSCLC patients
were profiled by miRNA array using a 3D-Gene Human miRNA Oligo
chips ver.20 (Toray Industries, Inc., Kamakura, Japan). Next, the
usefulness of selected miRNAs as biomarkers was clarified using
another 195 NSCLC cases. This study was approved by the
Institutional Review Board of the Ethics and Indications Committee
of Teikyo University (Tokyo, Japan). Written informed consent was
obtained from all patients. All patients underwent curative
surgery, and the median follow-up period was 20 months (range,
0.5–37 months). NSCLC patients were diagnosed according to the
current World Health Organization histological classification and
were staged according to the TNM classification of the
International Union Against Cancer (22,23).
Patients were treated with a standard regimen (uracil/tegafur for
TNM stage I patients and cisplatin plus vinorelbine for TNM stage
II and III patients) in accordance with the Guidelines of the Japan
Lung Cancer Society (24).
Post-operative follow-up was performed as follows: Confirmation of
recurrence in all patients was required to evaluate imaging or
pathological diagnosis. Physical examination and chest X-ray were
conducted every 3 months for 5 years, and computed tomography was
repeated every 6 months for 5 years after surgery.
Profiling study
For recurrence-specific miRNA microarray profiling,
6 cases of NSCLC (TNM stage III) were selected from the records of
the Department of Surgery, Teikyo University. Tumor recurrence was
found in 3 cases of the 6 cases after surgery. Recurrence sites
identified were the brain (2 cases) and the bones (1 case). As a
normal control, blood samples from 3 healthy volunteers were
examined. All blood samples were derived from patients who had not
received any chemotherapy or radiotherapy prior to surgery. Plasma
samples were separated from the blood, and exosomes were purified
from the plasma samples. Exosomal miRNA expression profiles were
examined using a 3D-Gene Human miRNA Oligo chips ver.20 (Toray
Industries, Inc., Kamakura, Japan). Details of the measuring
methods are described as follows.
Validation study
For the validation analysis, 195 consecutive NSCLC
patients and 30 healthy individuals treated between April 2012 and
July 2015 at Teikyo University Hospital (Tokyo, Japan) were
studied. These NSCLC patients included 129 cases with tumor-node
metastases (TNM) stage I, 34 cases with stage II and 32 cases with
stage III. All blood samples were derived from patients who had not
received any chemotherapy or radiotherapy prior to surgery. Plasma
samples were separated from the blood, and exosomes were purified
from the plasma samples. Exosomal miRNA expression was analyzed
using reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). Details of the measuring method are described as
follows.
Purification of exosomes from
plasma
A peripheral blood specimen (5 ml) was drawn from
each patient prior to surgery. Ethylenediaminetetraacetic acid was
used as an anticoagulant. Plasma and blood cells were separated by
centrifugation at 1,200 × g for 10 min at 4°C. Plasma (1.0
ml) was used for microarray analysis, and 500 µl was used for
RT-qPCR. The exosomes of the plasma were purified by the
ultracentrifugation method, as described previously (21). In brief, the exosomes were separated
by ultracentrifugation at 100,000 × g for 70 min at 4°C, and
then the pellets were washed with phosphate-buffered saline (PBS)
and stored at −80°C for microarray and RT-qPCR analysis.
Transmission electron microscopy of
exosomes
The isolated exosomes were dissolved in PBS, and a
drop of the suspension was placed on a carbon-coated copper grid
for 10 sec. The grid was then removed and excess liquid was drained
by filter paper. The grid was put in contact with a drop of 2%
uranyl acetate and phosphotungstic acid for 5 sec, and excess
liquid was then removed. The grid was allowed to dry for several
min and was then examined using an electron microscope (HITACHI
H-7600; Hitachi Ltd., Tokyo, Japan) at 100 kV.
miRNA isolation from exosomes
RNA was isolated from exosomes using the miRNeasy
serum/plasma kit (Qiagen, Hilden, Germany). Exosomes purified from
specific volumes of plasma (1.0 ml for microarray analysis and 500
µl for RT-qPCR assay) were diluted with 1 ml Qiazol Lysis Reagent.
Following incubation for 5 min, 3.5 µl of a spike-in control
(cel-miR-39 mimic) was added to each sample. Subsequent extraction
and cartridge work were performed according to the manufacturer's
protocols. The RNA quality was assessed using an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
miRNA microarray analysis
Exosomal miRNA expression profiles were examined
using 3D-Gene Human miRNA Oligo chips ver. 20 (Toray Industries,
Inc. Tokyo, Japan), according to the manufacturer's protocol.
Fluorescence signals were scanned and analyzed using the 3D-Gene
Scanner (Toray Industries, Inc.). A total of 2578 genes were
mounted on this chip. The raw data from each spot were normalized
by subtraction of the background signal mean intensity, determined
by the 95% confidence intervals of the signal intensities of all
blank spots. Valid measurements were considered those in which the
signal intensity of the duplicate spots was >2 standard
deviations of the background signal intensity.
RT-qPCR for miRNAs
Exosomal miRNA expression was assayed using RT-qPCR.
cDNA was synthesized from total RNA using Taqman microRNA primers
specific for hsa-miR-21 (assay ID 000397), has-miR-4257 (assay ID
244369) and has-miR-16a (assay ID 000391) (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and a TaqMan Micro-RNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.). miR-16a was
used as an internal control, as it is reported to be a reliable
endogenous control for miRNA analysis in RT-qPCR for humans
(21). RT-qPCR was performed using
Lightcycler-480 (Roche Applied Science, Basel, Switzerland) and
Taqman Universal PCR Master Mix (Thermo Fisher Scientific, Inc.)
following the manufacturer's protocols. Relative quantification of
miRNA expression was calculated using the 2−ΔΔCq method
(25). ΔCq was calculated by
subtracting the Cq values of miR-16 from the Cq values of miR-21 or
miR-4257. ΔΔCq was then calculated by subtracting the ΔCq of the
healthy control from the ΔCq of the sample. For healthy controls,
plasma exosome samples of 30 healthy volunteers were used.
Statistical analysis
The data are expressed as the mean ± standard
deviation. The correlations between microRNA expression and
clinicopathological factors were analyzed using Student's t-test,
the χ2 test and an analysis of variance. Disease-free
survival (DFS) curves were analyzed using the Kaplan-Meier survival
curve method, and the differences were examined using log-rank
tests. Cox proportional-hazards regression analysis was used to
estimate univariate and multivariate hazard ratios for DFS. All
P-values are two-sided, and P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
performed using the JMP 9.0 software (SAS Institute, Inc., Tokyo,
Japan).
Results
Identification of exosomes in
plasma
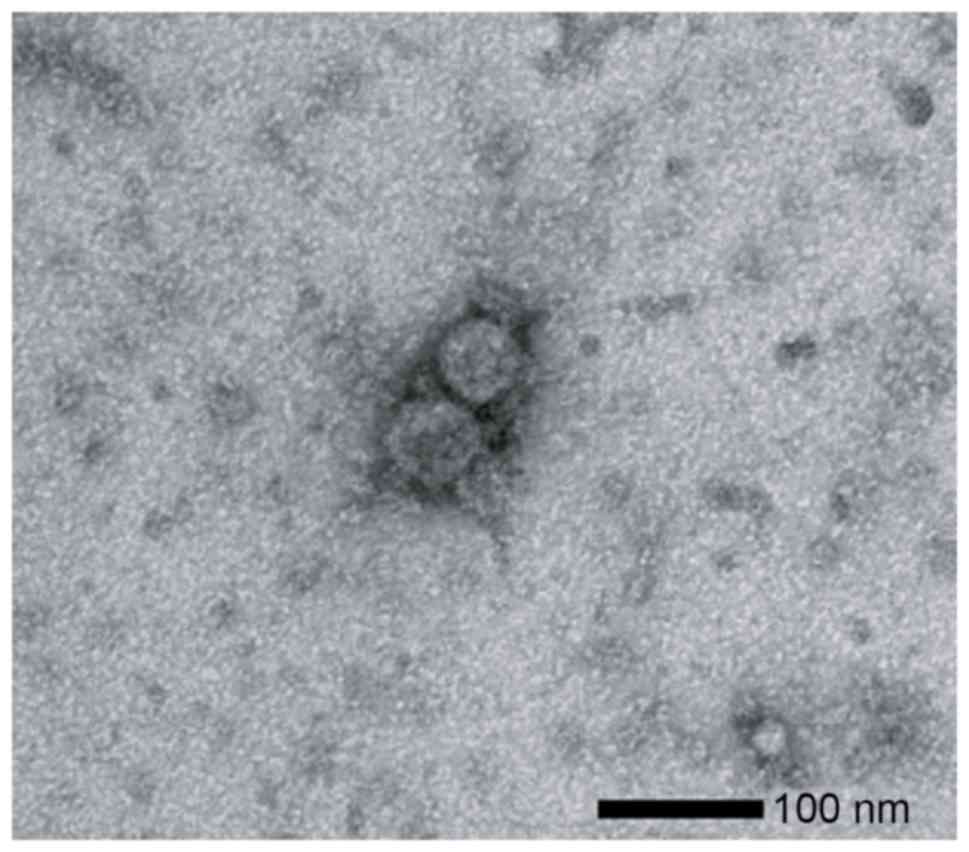
To verify the isolation of the exosomes from the
plasma, transmission electron microscopy results were examined of
ultracentrifugation samples from the NSCLC patients. Images of
round microvesicles with diameters of 50–100 nm were captured, as
shown in a representative sample in Fig.
1.
Exosomal miRNA profile of NSCLC
patients
To identify the recurrence-specific miRNA profiling
in the plasma exosomes of the NSCLC patients, the miRNA microarray
analyses of samples from 3 patients with recurrence after surgery,
3 patients without recurrence after surgery and 3 healthy
volunteers were examined. The clinical pathological characteristics
of the 6 NSCLC patients used for this analyses are shown in
Table I. In these patients, the
histological type and the TNM stages were the same in the
recurrence and recurrence-free cases. In the 3 recurrence cases,
the recurrent sites were the brain (2 cases) and the bones (1
case).
 | Table I.Clinicopathological factors of
patients used for the microRNA array. |
Table I.
Clinicopathological factors of
patients used for the microRNA array.
| Factors | Patients with
recurrence after surgery (n=3) | Patients without
recurrence after surgery (n=3) |
|---|
| Tumor size, cm |
|
|
|
<3 | 0 | 0 |
| ≥3 | 3 | 3 |
| Lymph node
metastasis |
|
|
| n
(−) | 0 | 0 |
| n
(+) | 3 | 3 |
| Lymphatic
invasion |
|
|
| Ly
(−) | 0 | 0 |
| Ly
(+) | 3 | 3 |
| Venous invasion |
|
|
| V
(−) | 0 | 0 |
| V
(+) | 3 | 3 |
| Histological
type |
|
|
|
Adenocarcinoma | 2 | 2 |
| Squamous
cell carcinoma | 1 | 1 |
| Tumor
differentiation |
|
|
| Well | 0 | 0 |
|
Moderate | 2 | 2 |
| Poor | 1 | 1 |
| TNM stage |
|
|
| I | 0 | 0 |
| II | 0 | 0 |
|
III | 3 | 3 |
Table II shows the
five most highly upregulated exosomal miRNAs in the patients with
recurrence. In this analysis, miR-4257 (miRBase no. MIMAT0016878)
showed the highest upregulation in the patients with recurrence
compared with those without recurrence and the healthy controls.
The mean fold-change of miR-4257 was 2.1 in comparison with that of
the recurrence-free patients, and 2.9 in comparison with that of
the healthy controls. miR-21 (miRBase no. MIMAT0000076) showed the
highest level after miR-4257. The mean fold-change of miR-21 was
1.7 in comparison with that of recurrence-free patients, and 2.5 in
comparison with that of the healthy controls. On the basis of these
profiling results, miR-4257 and miR-21 were selected as the
recurrence markers for NSCLC cases and were used in the present
study.
 | Table II.Five most highly upregulated miRNAs
in the non-small cell lung cancer patients with recurrence based on
the miRNA profiling array. |
Table II.
Five most highly upregulated miRNAs
in the non-small cell lung cancer patients with recurrence based on
the miRNA profiling array.
| Ranking | miRNA | miRBase no. | Fold-change
(comparison with recurrence-free patients) | Fold-change
(comparison with healthy controls) |
|---|
| 1 | miR-4257 | MIMAT0016878 | 2.1 | 2.9 |
| 2 | miR-21 | MIMAT0000076 | 1.7 | 2.5 |
| 3 | miR-887 | MIMAT0004951 | 1.4 | 1.9 |
| 4 | miR-6794 | MIMAT0027488 | 1.3 | 1.8 |
| 5 | miR-4476 | MIMAT0019003 | 1.3 | 1.7 |
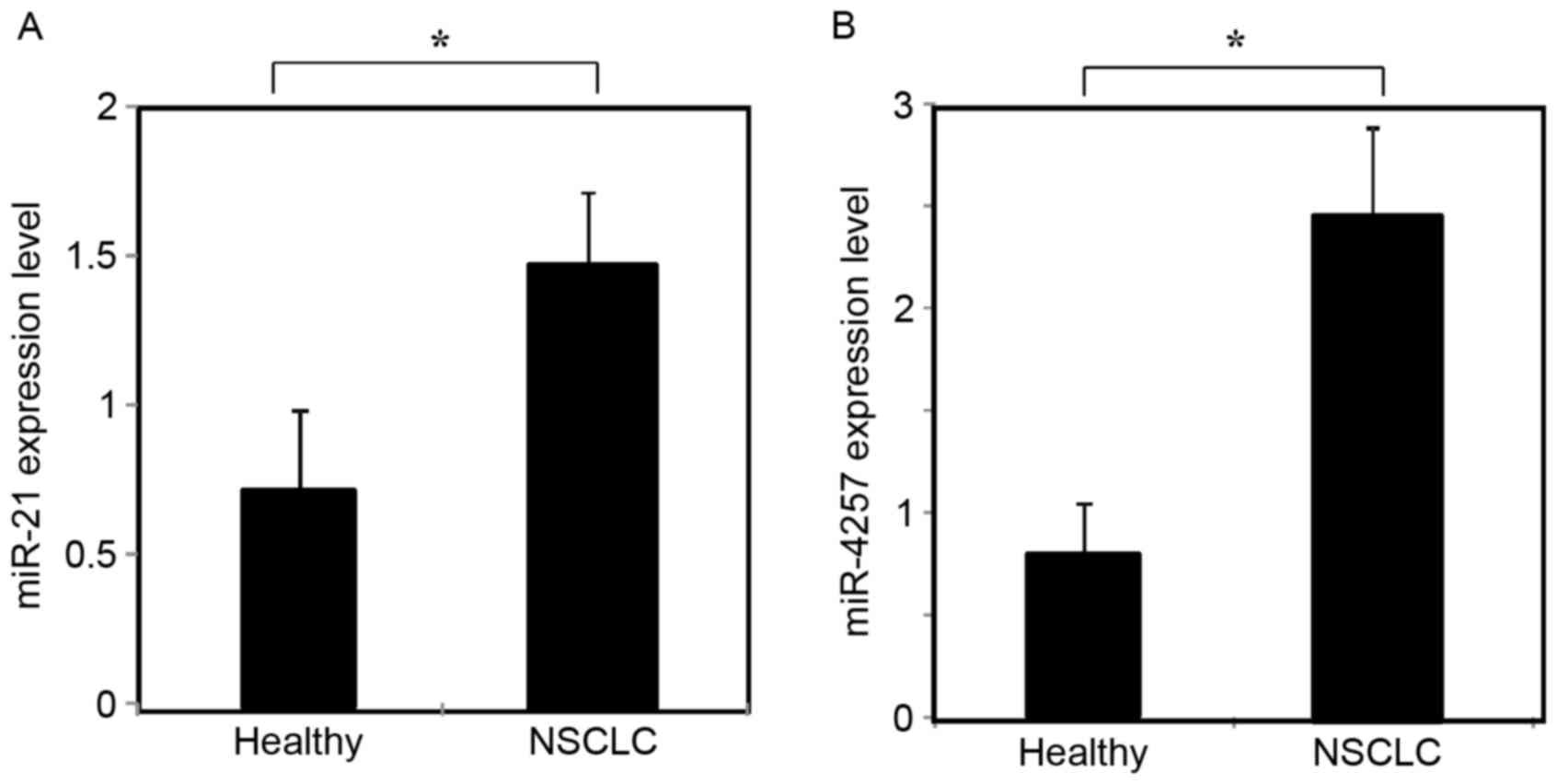
Expression of exosomal miR-21 and
miR-4257
The expression of miR-21 and miR-4257 was assessed
by RT-qPCR in the plasma exosomal samples from 195 patients with
NSCLC and 30 healthy volunteers. As shown in Fig. 2A and B, exosomal miR-21 and miR-4257
levels were significantly increased in NSCLC patients as compared
with those of healthy controls (P<0.01).
Clinicopathological significance of
miR-21 and miR-4257 expression in plasma exosomes
This study includes 195 NSCLC patients (112 men and
83 women), with a mean age of 71 years (range, 32–88 years). These
patients were drawn from a separate cohort to the miRNA microarray
study. To evaluate the correlation between the exosomal miR-21 and
miR-4257 expression levels and the clinicopathological
characteristics, patients were divided into two groups (high and
low). The cut-off level was determined as the average of the miR-21
and miR-4257 expression levels, as described previously (21). As shown in Table III, a statistically significant
association was observed between the high miR-21 group and tumor
size and TNM stage. A statistically significant association was
observed between the high miR-4257 group and lymphatic invasion,
histological type and TNM stage (Table
IV).
 | Table III.Association between
clinicopathological factors and high exosomal miR-21
expression. |
Table III.
Association between
clinicopathological factors and high exosomal miR-21
expression.
| Factors | No. of patients
(n=195) | miR-21 high
(n=88) | Positive rate,
% | P-value |
|---|
| Gender |
|
|
| 0.65 |
|
Male | 112 | 49 | 43.8 |
|
|
Female | 83 | 39 | 47.0 |
|
| Smoking status |
|
|
| 0.11 |
|
Non-smokers | 68 | 36 | 52.9 |
|
|
Smokers | 127 | 52 | 40.9 |
|
| Tumor size, cm |
|
|
| 0.02 |
|
<3 | 136 | 54 | 39.7 |
|
| ≥3 | 59 | 34 | 57.6 |
|
| Lymph node
metastasis |
|
|
| 0.10 |
| n
(−) | 148 | 59 | 39.9 |
|
| n
(+) | 47 | 29 | 61.7 |
|
| Lymphatic
invasion |
|
|
| 0.29 |
| Ly
(−) | 153 | 66 | 43.1 |
|
| Ly
(+) | 42 | 22 | 52.4 |
|
| Venous
invasion |
|
|
| 0.56 |
| V
(−) | 122 | 57 | 46.7 |
| V
(+) | 73 | 31 | 42.5 |
| Histological
type |
|
|
| 0.87 |
|
Adenocarcinoma | 134 | 60 | 44.8 |
|
Squamous cell carcinoma | 53 | 25 | 47.2 |
|
Others | 8 | 3 | 37.5 |
| Tumor
differentiation |
|
|
| 0.43 |
|
Well | 91 | 37 | 40.7 |
|
Moderate | 74 | 35 | 47.3 |
|
Poor | 30 | 16 | 53.3 |
| TNM stage |
|
|
| 0.03 |
| I,
II | 163 | 68 | 41.7 |
|
III | 32 | 20 | 62.5 |
 | Table IV.Association between
clinicopathological factors and high exosomal miR-4257
expression. |
Table IV.
Association between
clinicopathological factors and high exosomal miR-4257
expression.
| Factors | No. of patients
(n=195) | miR-4257 high
(n=54) | Positive rate,
% | P-value |
|---|
| Gender |
|
|
|
|
|
Male | 112 | 29 | 25.9 | 0.51 |
|
Female | 83 | 25 | 30.1 |
|
| Smoking status |
|
|
|
|
|
Non-smokers | 68 | 20 | 29.4 | 0.60 |
|
Smokers | 127 | 34 | 26.8 |
|
| Tumor size, cm |
|
|
|
|
|
<3 | 136 | 35 | 25.7 | 0.35 |
| ≥3 | 59 | 19 | 32.2 |
|
| Lymph node
metastasis |
|
|
|
|
| n
(−) | 148 | 36 | 24.3 | 0.06 |
| n
(+) | 47 | 18 | 38.3 |
|
| Lymphatic
invasion |
|
|
|
|
| Ly
(−) | 153 | 37 | 24.2 | 0.04 |
| Ly
(+) | 42 | 17 | 40.5 |
|
| Venous
invasion |
|
|
|
|
| V
(−) | 122 | 35 | 28.7 | 0.69 |
| V
(+) | 73 | 19 | 26.0 |
|
| Histological
type |
|
|
|
|
|
Adenocarcinoma | 134 | 47 | 35.1 | 0.02 |
|
Squamous cell carcinoma | 53 | 5 |
9.4 |
|
|
Others | 8 | 2 | 25.0 |
|
| Tumor
differentiation |
|
|
|
|
|
Well | 91 | 26 | 28.6 | 0.97 |
|
Moderate | 74 | 20 | 27.0 |
|
|
Poor | 30 | 8 | 26.7 |
|
| TNM stage |
|
|
|
|
| I,
II | 163 | 38 | 23.3 | <0.01 |
|
III | 32 | 16 | 50.0 |
|
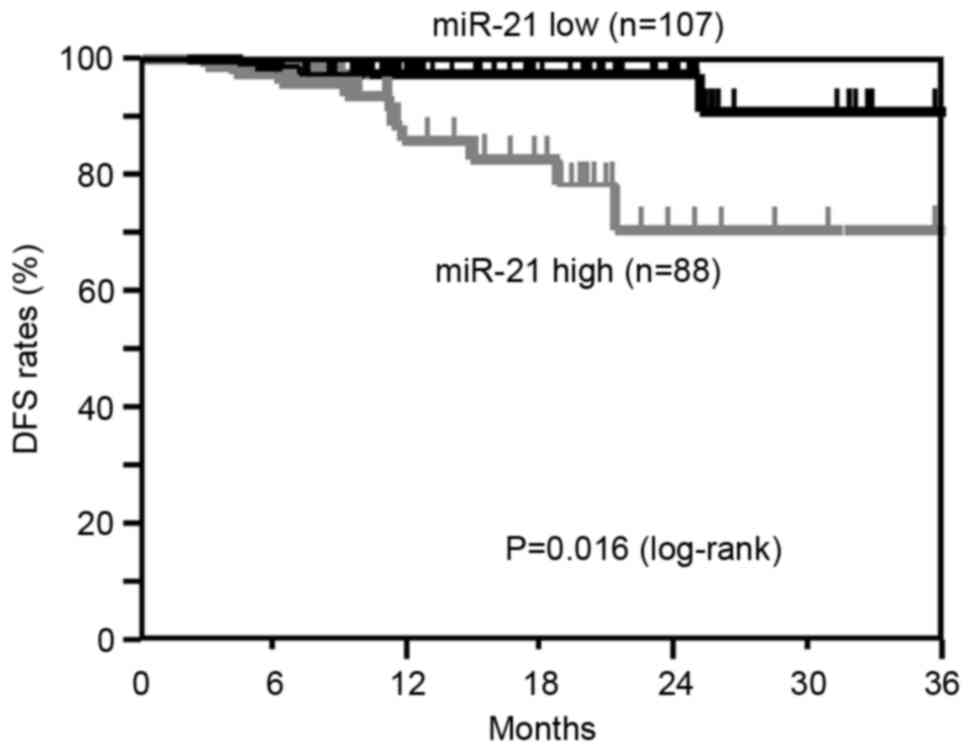
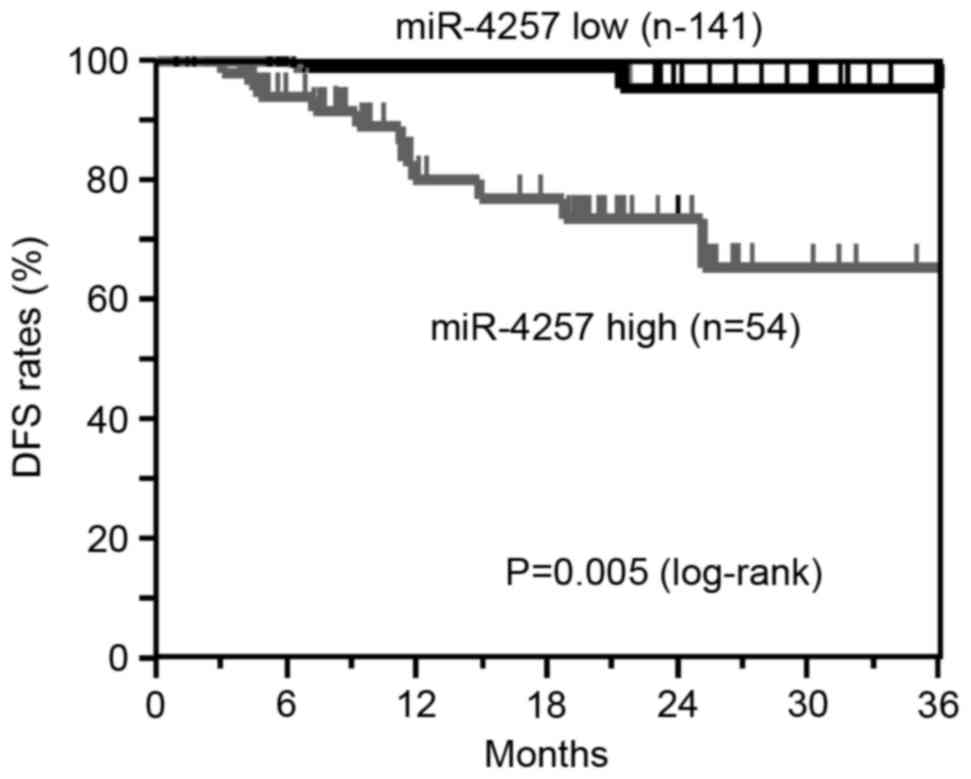
Correlation between miR-21 and
miR-4257 levels and DFS
The Kaplan-Meier DFS curves of the 195 NSCLC
patients (TNM stage I, 129 cases; TNM stage II, 34 cases; TNM stage
III, 32 cases) according to the status of miR-21 and miR-4257
levels were examined. Patients who had undergone curative surgery
were included in this analysis.
The DFS of patients in the high miR-21 group showed
a significantly worse survival rate than those in the low miR-21
group (log-rank, P=0.016; Fig. 3).
The DFS analysis revealed that patients in the high miR-4257 group
exhibited significantly worse survival rates than those in the low
miR-4257 group (log-rank, P=0.005; Fig.
4). These results suggest that the high expression of miR-21and
miR-4257 is associated with recurrence in NSCLC patients.
Univariate and multivariate Cox
analysis for DFS
Table V shows the
results of univariate and multivariate Cox proportional hazard
regression analysis for DFS in NSCLC patients (n=195). Multivariate
analysis was performed for all factors tested by univariate
analysis. In the univariate analysis, tumor size, lymphatic
invasion, TNM stage, and miR-21 and miR-4257 levels showed
significance for DFS. In the multivariate analysis, TNM stage, and
miR-21 and miR-4257 levels showed significance for DFS. These
results suggest that miR-21 and miR-4257 levels in tumor tissues
have an independent prognostic value for DFS in NSCLC patients.
 | Table V.Univariate and multivariate analyses
of the prognostic factors for disease-free survival in all
patients. |
Table V.
Univariate and multivariate analyses
of the prognostic factors for disease-free survival in all
patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Factors | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| Smoking status | 0.75 | 2.11
(0.64–9.43) |
0.229 |
0.05 | 1.05
(0.24–5.55) | 0.950 |
| Tumor size | 1.65 | 5.20
(1.73–17.27) |
0.004 |
0.76 | 2.13
(0.58–8.21) | 0.254 |
| Lymphatic
invasion | 1.29 | 3.63
(1.18–11.59) |
0.025 |
0.83 | 2.30
(0.71–7.65) | 0.163 |
| Venous
invasion | 0.98 | 2.66
(0.79–8.04) |
0.107 |
0.32 | 1.37
(0.24–6.89) | 0.708 |
| Histological
type | 0.73 | 2.07
(0.66–6.25) |
0.203 |
0.46 | 1.58
(0.31–7.88) | 0.576 |
| Tumor
differentiation | 0.83 | 2.29
(0.74–8.51) |
0.152 | −0.01 | 0.99
(0.25–4.55) | 0.991 |
| TNM stage | 2.59 | 13.35
(4.09–51.78) | <0.001 |
2.07 | 7.96
(1.97–36.09) | 0.004 |
| miR-21 | 1.37 | 3.93
(1.26–14.65) |
0.018 |
1.34 | 3.81
(1.13–14.93) | 0.031 |
| miR-4257 | 1.98 | 7.27
(2.21–32.49) |
0.001 |
2.09 | 8.09
(2.41–36.73) | 0.001 |
Discussion
The aim of the present study was to clarify the
specific plasma exosome miRNAs as biomarkers reflecting the
recurrence of NSCLC. It was found that plasma exosomal miR-21 and
miR-4257 could be potential biomarkers to predict recurrence in
NSCLC.
Recent studies showed that miRNAs are preserved in a
stable form in the exosome, are protected from endogenous RNase
activity and are important in the cell to cell information
processes (15,16). In the preliminary experiments of the
present study, the expression levels of miR-21 were compared
between the plasma and exosome samples separated from plasma in the
same NSCLC patients. It was found that the expression of exosomal
RNA was at high and stable levels as compared with that of the
plasma samples (data not shown). This may be due to the breakdown
of the miRNA in the plasma samples. However, the majority of the
previous studies in NSCLC did not examine the exosomal miRNAs
separated from the plasma or serum, which would likely include the
intact miRNA (7–13,26–29). In
the present study, microarray-based expression profiling of miRNAs
derived from exosomes in the plasma of 6 NSCLC patients with or
without recurrence was employed. In the microarray analysis,
miR-4257 and miR-21 showed marked upregulation in the NSCLC
patients with recurrence after surgery compared with those without
recurrence after surgery. Furthermore, these two exosomal miRNAs
levels were markedly increase as compared with those of the healthy
controls. miR-4257 showed the highest level of all miRNAs, with
miR21 showing the second highest level. From these results,
miR-4257 and miR-21 were selected as recurrence-specific biomarkers
for NSCLC patients.
It has been reported that serum miR-21 is
significantly associated with the promotion of tumor growth,
proliferation and progression, and the response of cancer to
chemotherapy (26). In an evaluation
of clinicopathological significance, Wang et al reported
that serum miR-21 expression was significantly correlated with
advanced TNM stage and the presence of lymph node metastasis in
NSCLC patients (27). However, the
status of plasma exosomal miR-21 expression and its potential as a
biomarker in NSCLC remains unclear. In the present study, using the
exosomal samples separated from the plasma of 195 NSCLC patients,
it was shown that high miR-21 expression has a significant
association with tumor size and TNM stage. By contrast, miR-4257 is
a novel microRNA and the association of clinicopathological factors
in NSCLC patients has not been hitherto reported. To the best of
our knowledge, the present study is the first to show that exosomal
miR-4257 levels are significantly associated with lymphatic
invasion, histological type and TNM stage.
The prognostic value of the miRNA of serum or plasma
samples as non-invasive biomarkers in NSCLC patients has been
reported. Sanfiorenzo et al showed the importance of the two
panels of plasma miRNAs for the prediction of recurrence in NSCLC
patients (28). The study reported
that one miRNA panel (high miR-155-5p, high miR-223-3p and low
miR-126a-3p) was significantly associated with a higher progression
risk in adenocarcinoma patients, and that another miRNA panel (high
miR-20a-5p, low miR-152-3p and low miR-199a-5p) was significant in
the prediction of likely survival rates of squamous cell carcinoma
patients. Gao et al reported that the miR-155 overexpression
in serum specimens may be a diagnostic marker for the early
detection of lung adenocarcinoma (29). The clinical significance of serum
miR-21 expression, but not exosomes, has also been reported. Liu
et al demonstrated that serum miR-21 and tumor miR-21,
miR-141 and miR-200c may be potential biomarkers for the diagnosis
of NSCLC (30). Wang et al
showed that serum miR-21 expression may be useful as a prognostic
marker for NSCLC patients (27).
Using a separate cohort of 195 NSCLC patients who
underwent curative surgery, the present study investigated the
prognostic significance of plasma exosomal miR-21 and miR-4257
levels using the Kaplan-Meier method and Cox multivariate analysis.
It was demonstrated that plasma exosomal miR-21 expression is a
potential predictive biomarker for recurrence in NSCLC patients.
Furthermore, the data showed that plasma exosomal miR-4257 also has
an independent prognostic impact on DFS in NSCLC patients. To the
best of our knowledge, this is the first study to clarify the
significant prognostic value of exosomal miR-4257 in NSCLC
patients.
miRNAs target protein-coding mRNAs at the
post-transcriptional level by directly cleaving the mRNA in two
different manners, namely, via directly cleaving the target mRNAs
and by inhibiting protein synthesis (31). Bioinformatically predicted targets for
miR-21 that have been experimentally validated include the tumor
suppressor genes programmed cell death 4 (PDCD4), phosphatase and
tensin homolog, tropomypsin 1 and reversion-inducing cysteine-rich
protein (32–34). PDCD4 has been characterized as a novel
tumor suppressor gene that acts as a suppressor of transformation,
progression, invasion and metalloproteinase activation, and as an
inducer of apoptosis. We have previously demonstrated a significant
inverse association between miR-21 and PDCD4 mRNA in colorectal
cancer patients (34). Through the
inhibition of the PDCD4 tumor suppressor gene by miR-21, tumor
growth may be promoted, leading to a poor prognosis. By contrast,
the target protein potential of miR-4257 has not been previously
reported. By searching the database, the ST13 tumor suppressor gene
was identified as a candidate target gene of miR-4257. The
expression of this gene is reported to be downregulated in
colorectal cancer tissue suggesting that it is a candidate tumor
suppressor gene (35). In the near
future, we aim to clarify the functional activity of miR-4257
through this target gene.
In conclusion, the present study demonstrates that
plasma exosomal miR-21 and miR-4257 show potential as biomarkers of
recurrence in NSCLC patients. These biomarkers may be therapeutic
targets in NSCLC.
Acknowledgements
The authors would like to thank Miss Junko Tamura
(Department of Surgery, Teikyo University School of Medicine) for
providing excellent technical assistance, and all members of the
respiratory surgery group of Teikyo University School of
Medicine.
Glossary
Abbreviations
Abbreviations:
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
NSCLC
|
non-small cell lung cancer
|
References
|
1
|
Wakao F, Nishimoto H, Katanoda K, et al:
Cancer Statistics in Japan 2015Editorial Board of the Cancer
Statistics in Japan (ed.). Foundation for Promotion of Cancer
Research c/o National Cancer Center; Tokyo: pp. 1–129. 2016
|
|
2
|
Maeda T, Ueda H, Tabata M, Kiura K,
Shibayama T, Gemba K, Takigawa N, Hiraki A, Katayama H and Harada
M: Prognostic factors in advanced non-small cell lung cancer:
Elevated serum levels of neuron specific enolase indicate poor
prognosis. Jpn J Cin Oncol. 30:534–541. 2000. View Article : Google Scholar
|
|
3
|
Del Vescovo V, Grasso M, Barbareschi M and
Denti MA: MicroRNAs as lung cancer biomarkers. World J Clin Oncol.
5:604–620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang QZ, Xu W, Habib N and Xu R: Potencial
uses of microRNA in lung cancer diagnosis, prognosis, and theray.
Curr Cnacer Drug Targets. 9:572–594. 2009. View Article : Google Scholar
|
|
5
|
Deng D, Liu Z and Du Y: Epigenetic
alterations as cancer diagnostic, prognostic, and predictive
biomatkers. Adv Genet. 71:125–176. 2010.PubMed/NCBI
|
|
6
|
Grady WM and Tewai M: The next thing in
prognostic molecular markers: MicroRNA signatures of cancer. Gut.
59:706–708. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen X, Ba Y, Ma I, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characteruzation of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Heegaard NH, Schetter AJ, Welsh JA, Yoneda
M, Bowman ED and Harris CC: Circulating micro-RNA expression
profiles in early stage nonsmall cell lung cancer. Int J Cancer.
130:1378–1386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ulivi P and Zoli W: miRNAs as non-invasive
biomarkers for lung cancer diagnosis. Molecules. 19:8220–8237.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Powrózek T, Krawczyk P, Kowalski DM,
Winiarczyk K, Olszyna-Serementa M and Milanowski J: Plasma
circulating microRNA-944 and microRNA-3662 as potential histologic
type-specific early lung cancer biomarkers. Transl Res.
166:315–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng D, Haddadin S, Wang Y, Gu LQ, Perry
MC, Freter CE and Wang MX: Plasma microRNAs as novel biomarker for
early detection of lung cancer. Int J Clin Exp Pathol. 4:575–586.
2011.PubMed/NCBI
|
|
12
|
Kowal J, Tkach M and Théry C: Biogenesis
and secretion of exosome. Curr Opin Cell Biol. 29:116–125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ge Q, Zhou Y, Lu J, Bai Y, Xie X and Lu Z:
miRNA in plasma exosome is stable under different storage
conditions. Molecules. 19:1567–1575. 2014. View Article : Google Scholar
|
|
14
|
Palma J, Yaddannapudi SC, Pigati L, Havens
MA, Jeong S, Weiner GA, Weimer KM, Stern B, Hastings ML and Duelli
DM: MicroRNAs are exported from malignant cells in customized
particles. Nucleic Acids Res. 40:9125–9138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Simons M and Raposo G: Exosomes-vesicular
carriers for intercelluar communication. Curr Opin Cell Biol.
21:575–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peinado H, Alečković M, Lavotshkin S,
Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M,
Williams C, García-Santos G, Ghajar C, et al: Melanoma exosomes
educate bone marrow progenitor cells toward a pro-metastatic
phenotype through MET. Nat Med. 18:883–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanisms of genetic exchange between cells.
Nat cell Bio. 9:654–659. 2007. View
Article : Google Scholar
|
|
18
|
Hannafon BN and Ding WQ: Intercellular
communication by exosome-derived microRNAs in cancer. Int J Mol
Sci. 14:14240–14269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanaka Y, Kamohara H, Kinoshita K,
Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M and Baba H:
Clinical impact of serum exosomal microRNA-21 as a clinical
biomarker in human esophageal squamous cell carcnoma. Cancer.
119:1159–1167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H and Sonoda
H: Circulatung exosomal microRNAs as biomarkers of colon cancer.
PLoS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sugimachi K, Matsumura T, Hirata H, Uchi
R, Ueda M, Ueo H, Shinden Y, Iguchi T, Eguchi H, Shirabe K, et al:
Idenification of a bona fide microRNA biomarker in serum exosomes
that predicts hepatocellular carcinoma recurrence after liver
transplantation. Br J Cancer. 112:532–538. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Travis WD, Brambilla E, Buke AP, Marx A
and Nicholson AG: WHO classification of tumors of the lung, pleura,
thymus and heartBosman FT, Jaffe ES, Lakhani SR and Ohgaki H:
International Agency for Research on Cancer; France: pp. pp1–pp412.
2015
|
|
23
|
General rule for clinical and pathological
record of lung cancer, . The Japan Lung Cancer Society (ed.).
Kenehara Co. Ltd; Tokyo: pp. pp1–pp213. 2014
|
|
24
|
Guidelines 2014 for treatment of lung
cancer by EBM, . Japan Lung Cancer Society (ed.). Kenehara Co. Ltd;
Tokyo: pp. pp1–pp201. 2014
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Li P, Ju H, Pesta M, Kulda V, Jin
W, Cai M, Liu C, Wu H, Xu J, et al: Diagnostic and prognostic value
of microRNA-21 in colorectal cancer: An original study and
individual participant data meta-analysis. Cancer Epidemiol
Biomarkers Prev. 23:2783–2792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang
KM and De W: Prognostic significance of serum miRNA-21 expression
in human non-small cell lung cancer. J Surg Oncol. 104:847–851.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sanfiorenzo C, Iiie MI, Belaid A, Barlési
F, Mouroux J, Marquette CH, Brest P and Hofman P: Two panels of
plasma microRNAs as non-invasive biomarkers for prediction of
recurrence in resectable NSCLC. PLoS One. 8:e545962013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao F, Chang J, Wang H and Ahang G:
Potential diagnostic value of miR-155 in serum from lung
adenocarcinoma patients. Oncol Rep. 31:351–357. 2014.PubMed/NCBI
|
|
30
|
Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ,
Wang YK, Zeng F, Zhou JH and Zhang YK: High expression of serum
miR-21 and tumor miR-200c associated with poor prognosis in
patients with lung cancer. Med Oncol. 29:618–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Motoyama K, Onoue H, Mimori K, Tanaka F,
Kojima K, Uetake H, Sugihara K and Mori M: Clinicopathological and
prognostic significance of pdcd4 and microRNA-21 in human gastric
cancer. Int J Oncol. 36:1089–1095. 2010.PubMed/NCBI
|
|
34
|
Horiuchi A, Iinuma H, Akahane T, Shimada R
and Watanabe T: Prognostic significance of PDCD4 expression and
association with microRNA-21 in each Dukes' stage of colorectal
cancer patients. Oncol Rep. 27:1384–1392. 2012.PubMed/NCBI
|
|
35
|
Bai R, Shi Z, Zhang JW, Li D, Zhu YL and
Zheng S: ST13, a proliferation regulator, inhibits growth and
migration of colorectal cancer cell lines. J Zhejiang Univ Sci B.
13:884–893. 2012. View Article : Google Scholar : PubMed/NCBI
|