Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide, and has multiple risk factors, associated
with lifestyle, genetics and the environment (1). Currently, the main therapeutic strategy
used to treat CRC is surgical resection, however its effectiveness
is limited in patients with locally invasive or metastatic disease.
Therefore, understanding the mechanisms of CRC disease progression
is essential in identifying potential biomarkers and improving CRC
patient prognosis and treatment.
MicroRNAs (miRNAs) are small non-protein coding RNAs
20–24 nucleotides long, that negatively regulate gene expression by
binding to the 3′ untranslated region (3′UTR) of corresponding
target messenger RNAs (mRNAs) (2).
Abnormal miRNA expression is thought to contribute to tumorigenesis
and carcinoma progression of various human cancers (3). Previous studies have demonstrated that
analysis of the genes targeted by miRNA may identify altered miRNA
regulatory networks involved in tumor pathogenesis; and miRNA
itself may be used to identify cancer-specific signatures for CRC
diagnosis (4). A number of studies
have demonstrated that downregulating the expression of tumor
suppressor miRNAs, such as miR-219-5p, may increase CRC development
and progression (5). miR-219-5p
inhibits papillary thyroid carcinoma cell growth and invasion by
targeting estrogen receptor 1 (6),
and targets glypican-3, inhibiting hepatocellular carcinoma cell
proliferation (7). Furthermore,
miR-219-5p regulates the receptor tyrosine kinase pathway in
glioblastoma by targeting the epidermal growth factor receptor
(8), and may regulate Sall4 in CRC
cells (5).
Calcyphosine, a calcium binding protein of 24 kDa,
was originally isolated from the canine thyroid cDNA library as a
major phosphorylated substrate for protein kinase A following
stimulation of thyroid cells by thyrotropin (8). As it is phosphorylated in a
cAMP-dependent manner, it is considered to be implicated in the
cross-signaling between these cascades to coordinate cellular
proliferation and differentiation (9). Calcyphosine is also expressed in the
brain, salivary glands and lungs of dogs (10). Moreover, calcyphosine functions as a
candidate for cross-talk between cAMP-mediated and inositol
trisphosphate/Ca2+ mediated signal transduction pathways
(11). Calcyphosine was thus
identified as a tumor-specific protein that may serve as a tumor
marker for a novel subgroup of ependymomas and as a potential drug
target for therapy in pediatric brain tumors (12). The present study aims to identify a
novel target gene of miR-219-5p and demonstrate that calcyphosin
protein (CAPS) is targeted by miR-219-5p in human CRC, to elucidate
the mechanism by which miR-219-5p suppresses CRC progression. This
would allow the clinical value of miR-219-5p in treating CRC
patients to be evaluated.
Materials and methods
Tissue specimens and cell culture
Fresh CRC tissue specimens, and specimens from
adjacent non-tumor tissues, were collected from 34 patients with
CRC, who received surgical treatment at the Affiliated Hospital of
Nantong University between May 2013 and May 2014. None of the
patients had received prior neoadjuvant treatment. Samples were
immediately frozen in liquid nitrogen, and stored at −80°C. All
human tissue was collected using protocols approved by the Ethics
Committee of Affliated Hospital of Nantong University.
Cell culture and transfection
Human ileocecal colorectal adenocarcinoma HCT-8
cells obtained from the Type Culture Collection of Chinese Academy
of Sciences (Shanghai, China) were maintained in RPMI-1640 Medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS, Invitrogen; Thermo
Fisher Scientific, Inc.) in a humidified incubator with a mixture
of 5% CO2 at 37°C. HCT-8 cells were then transfected
with a scrambled miR-219-5p inhibitor (Thermo Fisher Scientific,
Inc.), miR-219-5p mimics, or miRNA, which acted as a negative
control (NC). For CAPS functional analysis, the coding sequence of
CAPS was amplified and sub cloned into the pcDNA3.1 (+) vector
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer instructions, in order to induce overexpression of
endogenous CAPS. Following this, HCT-8 cells were transfected
either with an empty vector or a CAPS-expressing plasmid, using
lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using a Trizol®
Plus RNA purification kit (Invitrogen; Thermo Fisher Scientific,
Inc), and cDNA was synthesized from total RNA treated with DNase I
using an Omniscript reverse transcription kit (Qiagen Inc.,
Valencia, CA, USA) following the supplier's instructions. qPCR was
performed using the HotStart-IT SYBR Green qPCR Master Mix (2X; USB
Molecular Biology Reagents and Biochemicals; Thermo Fisher
Scientific Inc.) to detect mRNA levels in CRC cells. According to
the HotStart-IT protocol, 25 µl reactions were run with 2 µl cDNA.
PCR experiments were performed in a LightCycler 480 system (Roche
Diagnostics, Basel, Switzerland). The PCR procedure was as follows:
Hot start at 95°C for 10 min; 40 cycles of
amplification/quantification at 95°C for 10 sec, 60°C for 30 sec,
and 72°C for 30 sec during which time fluorescence was measured.
Melting curve analysis was performed using continuous fluorescence
acquisition from 65–97°C. According to the presence of a single
melt peak, these cycling parameters generated single amplicons for
both primer sets used. The sequences of the primers for CAPS were:
Forward, 5′-AGGCACCTTCCACTAGCAACAG-3′ and reverse,
5′-CCATGCTTGGTCTGGGCTCT-3′. The expression level of each target
gene was normalized by the Cq value of U6 non-coding small nuclear
RNA (U6), or glyceraldehyde 3-phosphate dehydrogenase (used as
internal controls) using the 2-ΔΔCt relative
quantification method (13).
For miRNA analysis, total RNA was extracted as
described in the previous paragraph. Reverse transcription was
performed using a miRNA 1st-Strand cDNA Synthesis kit (Stratagene;
Agilent Technologies, Inc., Santa Clara, CA, USA). The cDNA were
subjected to qPCR using the High-Specificity miRNA RT-qPCR
Detection kit (Stratagene; Agilent Technologies, Inc.). The primers
used were for hsa-miR-219-5p, and were as follows: Forward
5′-ACACTCCAGCTGGGTGATTGTCCAAACGC-3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′; and U6, forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. Finally, the relative
miR-219-5P level was normalized to the endogenous reference gene U6
for each sample in triplicate, and was calculated by the
2−ΔΔCq relative quantification method (13).
Western blot analysis
Protein from tumor tissue was extracted by lysis
buffer containing protease inhibitors. Wells were loaded with 10
µg/well of protein from each sample and then transferred to a
polyvinylidene fluoride membrane. Blocking was subsequently carried
out for 2 h using 5% non-fat milk in a mixture of Tris-buffered
saline and Tween 20 (TBST). Following incubation with the primary
monoclonal rabbit anti-human CAPS antibody (cat no. ab186741,
1:1,000, Abcam, Cambridge, MA, USA) overnight at 4°C, the membranes
were washed with TBST three times for 10 min, and then incubated
with horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (cat no. ab97051, 1:5,000, Abcam) for 12 h at 4°C. The ECL
western blotting detection reagent (GE healthcare life sciences,
Chalfont, UK) was used to detect signals prior to film development.
β-actin was used as the loading control.
MTT assay
The MTT assay was used to determine the effect of
miR-219-5p on cellular proliferation. A total of 1×103 cells
containing 200 µl RPMI-1640 supplemented with 10% FBS were plated
in each well of a 96-well plate. Following 1, 2, 3 days of
incubation, 20 µl MTT (5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA)
was added, followed by 4 h incubation at 37°C in a 5%
CO2 incubator. The supernatant was subsequently removed
and 150 µl dimethyl sulfoxide was added. To dissolve MTT crystals,
all plates were shaken for 10 min at room temperature. The
absorbance values of each sample were read at 490 nm using a micro
plate reader (Model 550, Bio-Rad Laboratories, Shanghai, China).
Each experiment was repeated ≤3x.
Colony formation assay
Following transfection of miR-219-5p mimics (50
nmol/l), miR-219-5p inhibitor (50 nmol/l) or scramble (NC; 50
nmol/l), HCT-8 cells were plated in a 24-well plate for 24 h. The
cells were then collected and seeded (1,000 cells/well) in a fresh
6-well plate for 14 days. Surviving colonies (>50 cells per
colony) were counted following fixation with 1:1 methanol and
acetone, and staining with 5% Gentian Violet (ICM Pharma Pte. Ltd.,
Singapore, Singapore). The experiment was carried out in
triplicate.
Transwell assay
A transwell assay (BD Biosciences, Franklin Lakes,
NJ, USA) was used to perform cell invasion according to the
manufacturer's instructions. Mimics and inhibitors of miR-219-5p
transfected cells were harvested 24 h following transfection.
2.0×105 transfected cells or untreated cells in serum-free medium
were then added to each upper insert pre-coated with Matrigel
matrix, and 600 µl 10% FBS medium was added to the matched lower
chamber. Following incubation, non-invasive cells were removed from
the upper surface of the Transwell membrane by a cotton swab.
Invasive cells on the lower membrane surface were fixed with
methanol, stained with 0.1% crystal violet, photographed, and
counted.
Dual-luciferase reporter assay
HCT-8 cells were seeded in 24-well plates (1×105
cells/well) and incubated for 24 h prior to transfection. For the
reporter gene assay, the cells were co-transfected with either 0.5
µg pGL3-CAPS-3′UTR wild-type or pGL3-CAPS-3′UTR mutant plasmid,
0.05 ng pRL-TK control vector (Promega Corporation, Madison, WI,
USA), and 50 nmol/l either miR-219-5p mimic or scramble, using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
The firefly and renilla luciferase activities were measured
consecutively through a dual luciferase assay (Promega Corporation)
24 h after transfection. Luciferase intensity was measured using
the Dual Luciferase Reporter assay system (Promega Corporation,)
according to the manufacturer instructions.
Statistical analysis
All statistical analyses were performed using SPSS
software ver. 17.0 (SPSS, Inc., Chicago, IL, USA). Results were
compared using either Student's t-test or χ2 test. One-way analysis
of variance (ANOVA) test was used to analyze the significance of
differences between groups of various differentiations. All data
were presented as mean ± standard deviation (SD). P<0.05 was
considered to indicate a statistically significant difference.
Results
Over-expression of miR-219-5p inhibits
proliferation of CRC cells in vitro
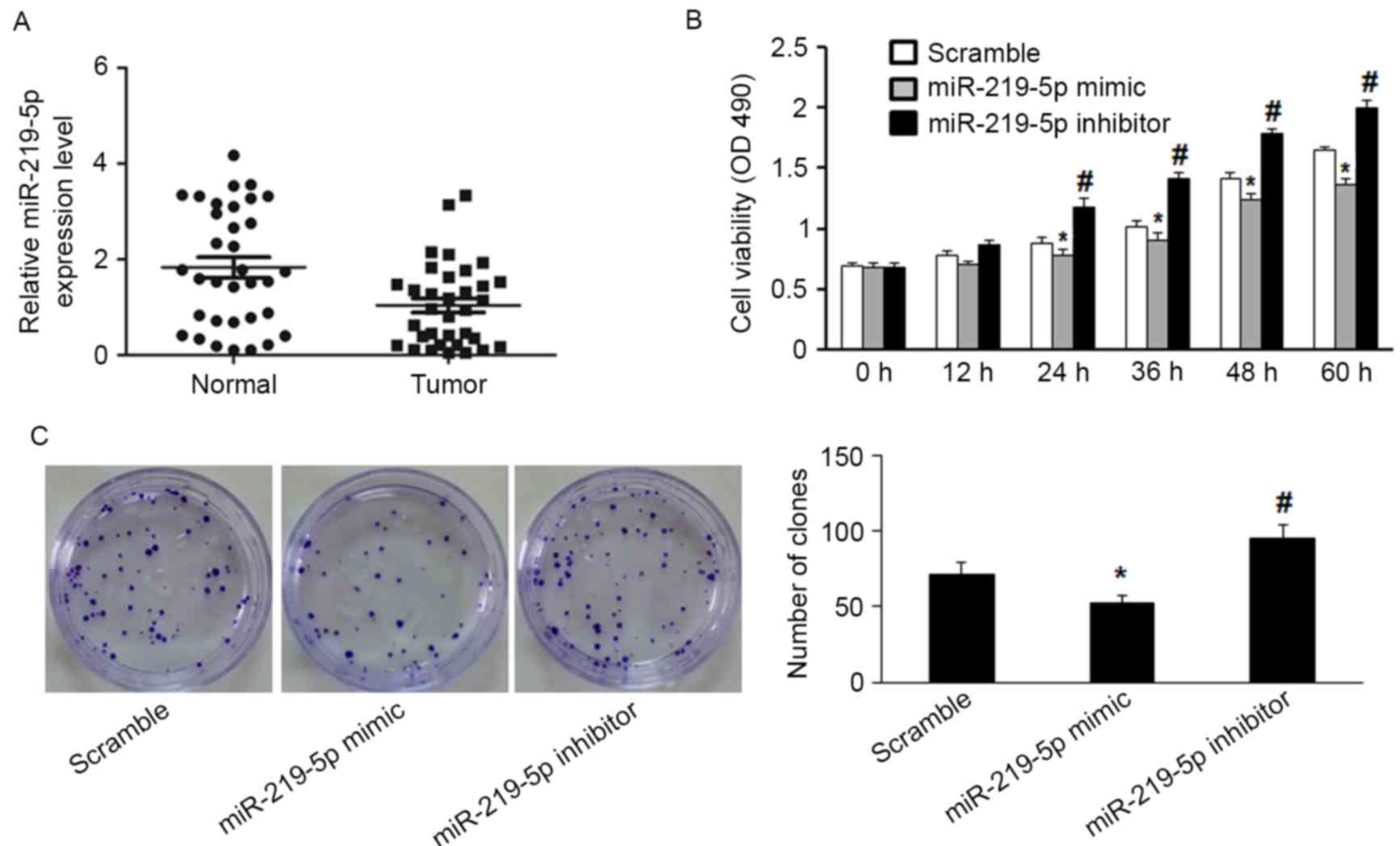
RT-qPCR detected significantly decreased miR-219-5p
expression in CRC tissue compared with the adjacent non-tumor
tissue (P=0.003, Fig. 1A). Moreover,
the functional role of miR-219-5p in CRC cells was evaluated by
measuring proliferation of HCT-8 cells. miR-219-5p expression was
upregulated following transfection with miR-219-5p mimic, and
downregulated by transfection with miR-219-5p inhibitor. The MTT
assay was used to evaluate HCT-8 cell proliferation. Transfection
with miR-219-5p mimics suppressed cell proliferation while
transfection with miR-219-5p inhibitor promoted cell proliferation
(Fig. 1B). The inhibitory effect of
miR-219-5p on the growth of CRC cells was confirmed by the colony
formation assay; transfection with miR-219-5p mimic led to a
significant reduction of colony numbers of HCT-8 cells compared
with transfection with scramble (Fig.
1C, P<0.05). HCT-8 cells transfected with miR-219-5p
inhibitor exhibited significantly increased cell proliferation
compared with the control group (Fig.
1C, P<0.05). These results demonstrate that miR-219-5p may
function as a tumor suppressor in CRC carcinogenesis.
CAPS-3′UTR is a potential functional
target of miR-219-5p
Previous studies have demonstrated that miR-219-5p
suppresses the expression of several genes, such as Sall4, by
targeting their 3′-UTR in CRC (5). To
identify new potential target genes of miR-219-5p, the websites
TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/) were used. Bioinformatic
analysis demonstrated that miR-219-5p directly targets the CAPS
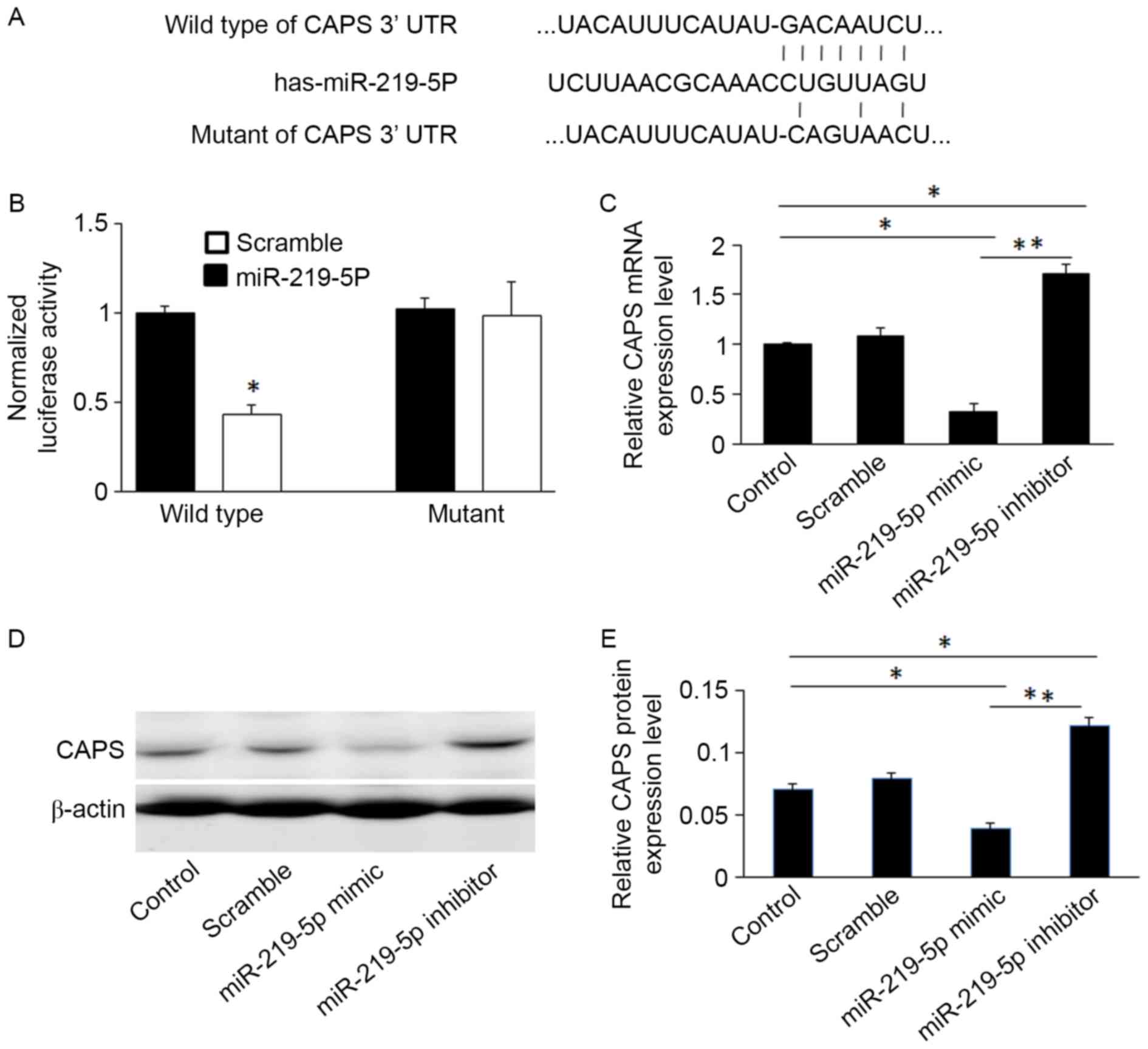
gene. To validate this, the wild-type and mutant sequences of the
CAPS 3′UTR were cloned into luciferase reporter vectors (sequences
shown in Fig. 2A). The luciferase
assay revealed that miR-219-5p significantly suppressed luciferase
activity of CAPS containing a wild-type 3′UTR, however it did not
suppress the activity of CAPS containing a mutant 3′UTR (P<0.05,
Fig. 2B). This confirms that
miR-219-5p targets the 3′UTR of CAPS.
miR-219-5p mimics significantly decreased CAPS
protein and mRNA levels compared to controls, whereas miR-219-5p
inhibitors increased levels of CAPS mRNA and protein in HCT-8 cells
(Figs. 2C and E). This was confirmed
by western blot analysis (Fig.
2D).
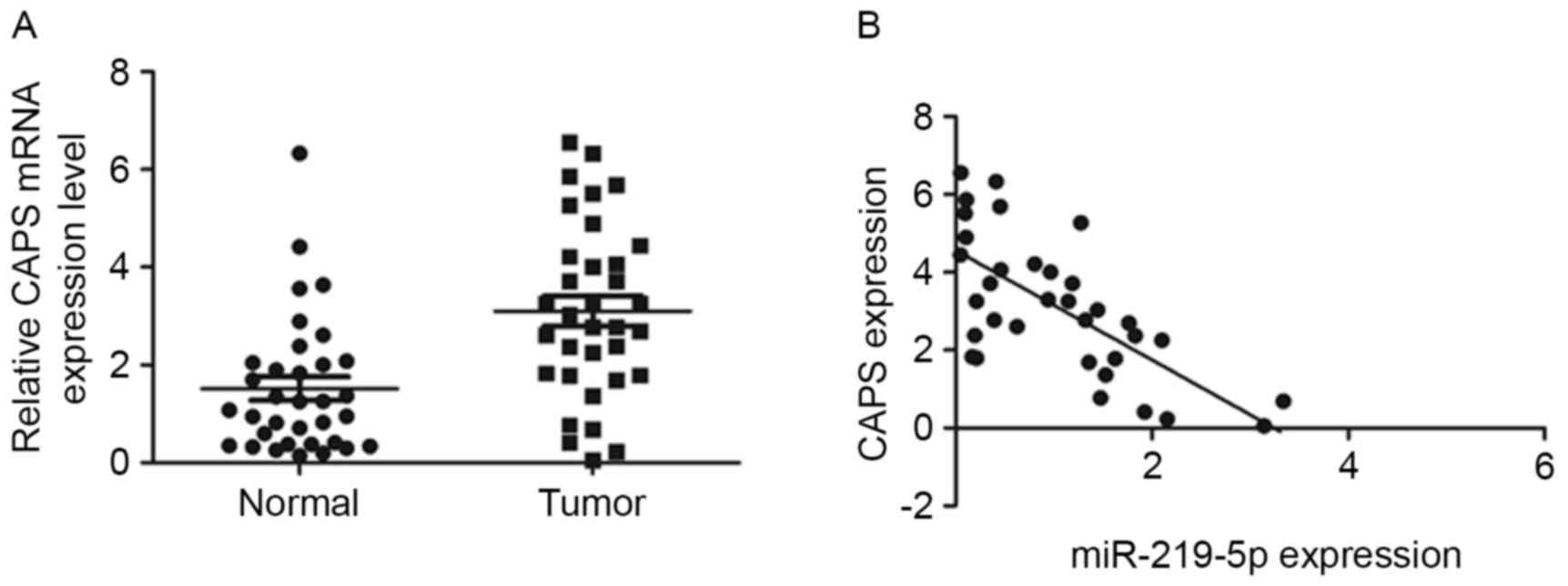
Furthermore, the mRNA level of CAPS in the 34 pairs
of tumor and adjacent non-tumor tissue were analyzed by RT-qPCR.
CAPS mRNA levels were significantly higher in CRC tissue than in
healthy tissue (P<0.001, Fig. 3A).
Furthermore, a significant negative correlation was observed
between miR-219-5p and CAPS expression in CRC samples
(r2=−0.47, P<0.001, Fig.
3B). Taken all together, the results of the current study
suggest that miR-219-5p decreases the expression of CAPS in CRC by
directly targeting CAPS 3′UTR.
Restoration of miR-219-5p inhibits
CAPS-mediated CRC cell invasion
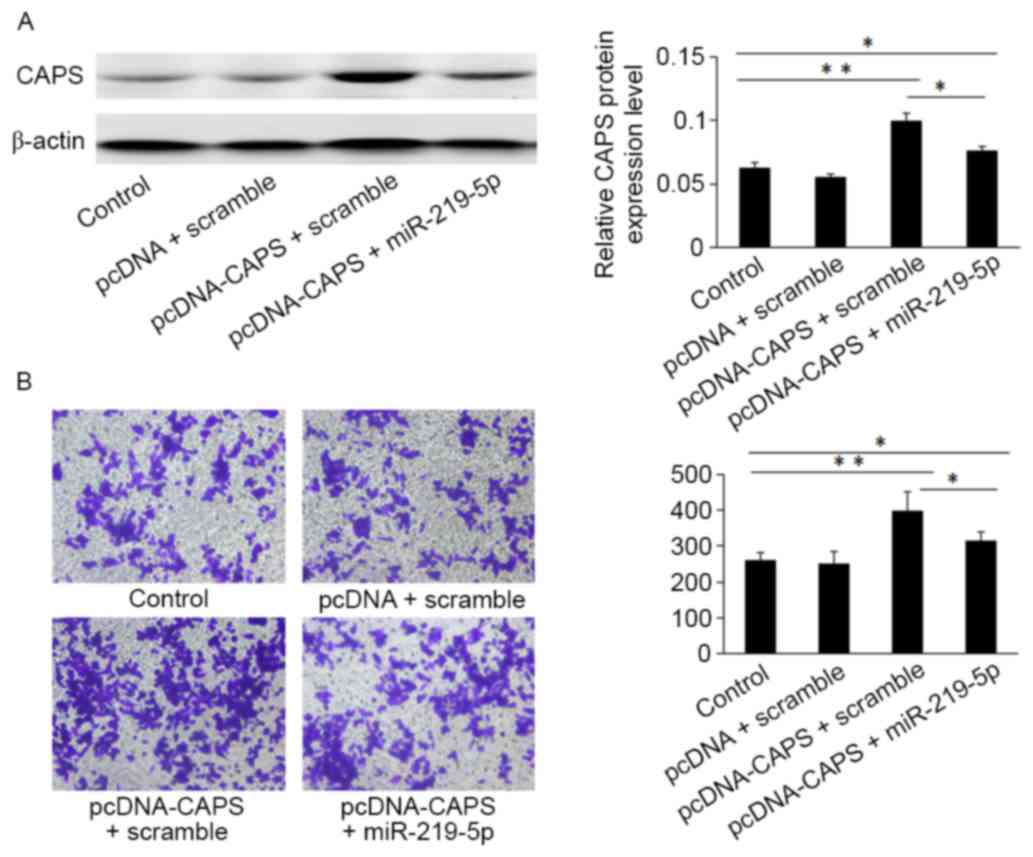
Western blot analysis indicated that HCT-8 cells
transfected with the pcDNA3.1 (+)-CAPS may enhance CAPS protein
expression compared with cells transfected with an empty vector
control (Fig. 4A). Furthermore, the
Transwell assay demonstrated that HCT-8 cells transfected with
miR-219-5p mimic and pcDNA3.1 (+)-CAPS overexpression of miR-219-5p
(312±12.46) exhibited significantly decreased migratory capacity
compared with CAPS-induced CRC cells transfected with CAPS and a
scramble (403±18.53; Fig. 4B,
P<0.05).
Discussion
Recently, the role of miRNAs in tumorigenesis and
their regulatory function in a number of biological processes
associated with cancer has been investigated. In human cancer,
miRNAs are usually located in genomic breakpoint regions and
function as tumor suppressor genes or oncogenes during tumor
development and progression (14).
miR-219-5p may act as a tumor manipulator in various types of
cancers by targeting downstream oncogenic molecules, thus
attenuating the malignant features of hepatocellular carcinoma
(7), papillary thyroid carcinoma
(6), glioblastoma (15) and colorectal cancer (5).
In the present study, analysis by RT-qPCR
demonstrated that miR-219-5p is downregulated in CRC tissue
compared with adjacent non-tumor tissues. Evidence gathered from
numerous studies highlights the significance of miRNAs as being
vital in the regulation of numerous physiological processes such as
cell proliferation, differentiation and apoptosis (16–18). Both
the MTT and colony formation assays demonstrated that miR-219-5p
significantly suppressed the growth and proliferation of CRC cells
in vitro. By contrast, downregulating miR-219-5p expression
promoted CRC cell proliferation.
Numerous studies have indicated that miRNAs are
deregulated in CRC, and the importance of this deregulation is
highlighted by the importance of the target genes they regulate
(19–21). The current study sought to identify
other potential targets of miR-219-5p to further understand how
miR-219-5p suppresses tumors in CRC. Target prediction algorithms
identified the binding sites for miR-219-5p in the 3′-UTR of
CAPS.
CAPS belongs to the calmodulin superfamily of
calcium-binding proteins and is regulated by cyclic AMP through
protein phosphorylation (22). It is
phosphorylated in a cAMP-dependent manner and thus may be
implicated in the cross-signaling between these cascades to
coordinate cell proliferation and differentiation (9), suggesting its involvment in
carcinogenesis. It has been proven that CAPS is expressed in many
tumors; overexpression of CAPS occurs in ependymoma (23), lung cancer (24), ovarian cancer (25) and endometrial cancer (26).
In conclusion, the present study identified CAPS as
a direct target of miR-219-5p in CRC cells. CAPS was upregulated in
CRC tissues, and miR-219-5p expression in CRC tissues was inversely
correlated with CAPS expression: Increasing miR-219-5p expression
downregulated the expression of CAPS protein, while knockdown of
miR-219-5p increased CAPS protein levels. Furthermore, CAPS-induced
cell invasion was reversed by miR-219-5p expression. Therefore,
miR-219-5p may function as a tumor suppressor and inhibit tumor
proliferation and invasion by regulating CAPS expression.
Acknowledgements
The present study was supported by the Affiliated
Hospital of Nantong University (grant no. TDFzh201413).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun Y, Wang L, Guo SC, Wu XB and Xu XH:
High-throughput sequencing to identify miRNA biomarkers in
colorectal cancer patients. Oncol Lett. 8:711–713. 2014.PubMed/NCBI
|
|
5
|
Cheng J, Deng R, Zhang P, Wu C, Wu K, Shi
L, Liu X, Bai J, Deng M, Shuai X, et al: miR-219-5p plays a tumor
suppressive role in colon cancer by targeting oncogene Sall4. Oncol
Rep. 34:1923–1932. 2015.PubMed/NCBI
|
|
6
|
Huang C, Cai Z, Huang M, Mao C, Zhang Q,
Lin Y, Zhang X, Tang B, Chen Y, Wang X, et al: miR-219-5p modulates
cell growth of papillary thyroid carcinoma by targeting estrogen
receptor α. J Clin Endocrinol Metab. 100:E204–E213. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang N, Lin J, Ruan J, Su N, Qing R, Liu
F, He B, Lv C, Zheng D and Luo R: MiR-219-5p inhibits
hepatocellular carcinoma cell proliferation by targeting
glypican-3. FEBS Lett. 586:884–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lecocq R, Lamy F and Dumont JE: Pattern of
protein phosphorylation in intact stimulated cells: Thyrotropin and
dog thyroid. Eur J Biochem. 102:147–152. 1980. View Article : Google Scholar
|
|
9
|
Clément S, Dumont JE and Schurmans S: Loss
of calcyphosine gene expression in mouse and other rodents. Biochem
Biophys Res Commun. 232:407–413. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lefort A, Lecocq R, Libert F, Lamy F,
Swillens S, Vassart G and Dumont JE: Cloning and sequencing of a
calcium-binding protein regulated by cyclic AMP in the thyroid.
Embo J. 8:111–116. 1989.PubMed/NCBI
|
|
11
|
Nemoto Y, Ikeda J, Katoh K, Koshimoto H,
Yoshihara Y and Mori K: R2D5 antigen: A calcium-binding
phosphoprotein predominantly expressed in olfactory receptor
neurons. J Cell Biol. 123:963–976. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong H, Li X, Lou Z, Xu X, Su D, Zhou X,
Zhou W, Bartlam M and Rao Z: Crystal-structure and biochemical
characterization of recombinant human calcyphosine delineates a
novel EF-hand-containing protein family. J Mol Biol. 383:455–464.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in Cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Y, Yin L, Jing H and Zhang H:
MicroRNA-219-5p exerts tumor suppressor function by targeting ROBO1
in glioblastoma. Tumour Biol. 36:8943–8951. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sen R, Ghosal S, Das S, Balti S and
Chakrabarti J: Competing endogenous RNA: The key to
posttranscriptional regulation. ScientificWorldJournal.
2014:8962062014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs-the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Esquela-kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma Y, Zhang P, Wang F, Zhang H, Yang J,
Peng J, Liu W and Qin H: miR-150 as a potential biomarker
associated with prognosis and therapeutic outcome in colorectal
cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schee K and Ø and Flatmark K Fodstad:
MicroRNAs as biomarkers in colorectal cancer. Am J Pathol.
177:1592–1599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He X, Dong Y, Wu CW, Zhao Z, Ng SS, Chan
FK, Sung JJ and Yu J: MicroRNA-218 inhibits cell cycle progression
and promotes apoptosis in colon cancer by downregulating BMI1
polycomb ring finger oncogene. Mol Med. 18:1491–1498.
2013.PubMed/NCBI
|
|
22
|
Kretsinger RH: Crystallographic studies of
calmodulin and homologs. Ann N Y Acad Sci. 356:14–19. 1980.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Bont JM, de Boer ML, Kros JM, Passier
MM, Reddingius RE, Smitt PA, Luider TM and Pieters R:
Identification of novel biomarkers in pediatric primitive
neuroectodermal tumors and ependymomas by proteome-wide analysis. J
Neuropathol Exp Neurol. 66:505–516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pastor MD, Nogal A, Molina-Pinelo S,
Meléndez R, Salinas A, De la Peña M González, Martín-Juan J, Corral
J, García-Carbonero R, Carnero A and Paz-Ares L: Identification of
proteomic signatures associated with lung cancer and COPD. J
Proteomics. 89:227–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Partheen K, Levan K, Osterberg L and
Horvath G: Expression analysis of stage III serous ovarian
adenocarcinoma distinguishes a sub-group of survivors. Eur J
Cancer. 42:2846–2854. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Min W, Huang C, Bai S, Tang M and
Zhao X: Proteomics-based approach identified differentially
expressed proteins with potential roles in endometrial carcinoma.
Int J Gynecol Cancer. 20:9–15. 2010. View Article : Google Scholar : PubMed/NCBI
|