Introduction
Glioma is the most common brain tumor and markedly
affects patient survival due to the high metastasis and recurrence
rate (1,2). According to the World Health
Organization (WHO), there are 4 malignancy grades, consisting of
grade I, which may develop in a benign pattern; grades II–III,
which may invade the adjacent brain tissues and gradually develop
into highly aggressive grade IV glioma, which is also termed
glioblastoma (3,4). Despite developments in therapies that
involve surgical resection, radiation therapy and chemotherapy, the
prognosis of glioma has not been significantly improved over the
past few decades (5). The overall
survival rate of high-grade gliomas is only 40% at 1 year, and the
5-year survival rate is <10% (6).
Thus, understanding the molecular etiology of glioma may aid the
development of more effective treatments.
Sineoculis homeobox homolog 1 (Six1) is a mammalian
homolog of the Drosophila sine oculis gene, and the gene is
highly conserved between Drosophila and humans (7,8). The
correct expression of this gene is crucial for the development of
multiple organs, including the brain, eye, ear, craniofacial
structures and kidney sensory structures (9–11). In
addition to the involvement of Six1 in the early development of
organs, the gene is often misexpressed in diverse tumors, including
breast cancer (12), ovarian cancer
(13,14), cervical cancer (15,16),
Wilms' tumors (17),
rhabdomyosarcomas (18) and
hepatocellular carcinoma (19).
Additionally, the misexpression of Six1 in cancer may induce
developmental programs out of context, contributing to tumor onset
and progression (20,21).
However, the association between Six1 expression and
glioma remains unknown. The present study aimed to investigate the
expression of Six1 in gliomas with distinct clinicopathological
features and to analyze the effect on the prognosis of glioma
patients.
Materials and methods
Patients and tissues
The present study enrolled 163 patients with glioma,
who had been clinically and histopathologically diagnosed and were
retrieved for tissue microarray (TMA) construction and
immunohistochemical (IHC) analysis from the Department of
Neurosurgery of Inner Mongolia People's Hospital (Hohhot, Inner
Mongolia, China). In accordance with the WHO classification, all
cases were classified as shown in Table
II, with 44 patients diagnosed with grade I disease, 67
patients diagnosed with grade II disease, 21 patients diagnosed
with grade III disease and 31 patients diagnosed with grade IV
disease. The mean age of patients at diagnosis was 45.26±10.43
years (range, 9–70 years), with 99 male and 64 female patients.
Follow-up data were available for 153 patients (range, 4–77 months;
mean, 40.9±19.95 months). The study protocol was performed with
approval from the Ethics Committee of the Inner Mongolia People's
Hospital, and informed consent was obtained from all patients.
 | Table II.Association between Six1 expression
and clinical and pathological factors in 163 patients with
glioma. |
Table II.
Association between Six1 expression
and clinical and pathological factors in 163 patients with
glioma.
|
|
| Six1 expression,
n |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | Total, n | Low | High | χ2 | P-value |
|---|
| Age |
|
|
|
1.978 |
0.160 |
| ≤45
years | 81 | 67 | 11 |
|
|
| >45
years | 82 | 74 | 8 |
|
|
| Gender |
|
|
|
0.777 |
0.678 |
|
Male | 99 | 85 | 14 |
|
|
|
Female | 64 | 56 | 8 |
|
|
| WHO grade |
|
|
| 29.622 | <0.001 |
| I | 44 | 42 | 2 |
|
|
| II | 67 | 63 | 4 |
|
|
|
III | 21 | 18 | 3 |
|
|
| IV | 31 | 18 | 13 |
|
|
TMA construction and
immunohistochemistry (IHC)
Representative sections of glioma or normal brain
tissues in the pre-existing paraffin-embedded tissue blocks were
determined according to the overlaid hematoxylin and eosin staining
slides. The TMA was constructed by excising a 1.0 mm diameter
cylinder from the representative section of each block using a
needle, and placing the cylinders into an array on a recipient
paraffin block. Subsequently, multiple 5-µm thick sections were cut
from the TMA block and mounted on microscope slides for IHC
analysis. The TMA consisted of a total of 163 cases of glioma and
16 cases of normal control paraffin-embedded tissue. The clinical
characteristics of the patients are summarized in Table I. The TMA slide was dried overnight at
37°C, deparaffinized in xylene, rehydrated in graded alcohol
solutions, and then immersed in 3% hydrogen peroxide for 10 min to
inactivate peroxidase activity. Antigen retrieval was performed by
microwave heating with 0.01 mol/l citrate buffer at 100°C for 15
min, and the slides were then cooled for 30 min at room temperature
to expose antigenic epitopes. The slides were pre-incubated with 5%
normal goat serum (Jetway Biotech, Co., Ltd., Guangzhou, China) at
room temperature for 30 min to reduce the non-specific reaction.
The primary rabbit anti-Six1 polyclonal antibody (cat. no.
HPA001893; Atlas Antibodies, Stockholm, Sweden) was diluted
(1:1,500) with 1X phosphate-buffered saline (PBS) and applied
overnight in a humidity chamber at 4°C. The slide was sequentially
incubated with a polymer peroxidase-labeled secondary antibody
(1:1,500; ZDR-5306; ZSGB-Bio, Beijing, China) for 30 min at room
temperature, and then visualized by catalysis of
3,3′-Diaminobenzidine Horseradish Peroxidase Color Development kit
(Beyotime Institute of Biotechnology, Haimen, China). Finally, the
sections were counterstained by hematoxylin. A known IHC-positive
slide was used as a positive control, and PBS replaced the
anti-Six1 primary antibody in the condition that was used as a
control.
 | Table I.Status of Six1 expression in all 163
glioma tissues and 16 normal brain tissues. |
Table I.
Status of Six1 expression in all 163
glioma tissues and 16 normal brain tissues.
|
|
| Six1 expression
status, n |
|
|---|
|
|
|
|
|
|---|
| Tissue type | Total, n | − | + | ++ | +++ | Percentage, % |
|---|
| Grade I | 44 | 35 | 7 | 1 | 1 |
4.55 |
| Grade II | 67 | 38 | 25 | 3 | 1 |
5.97 |
| Grade III | 21 | 5 | 13 | 2 | 1 | 14.29 |
| Grade IV | 31 | 5 | 13 | 9 | 4 | 41.94 |
| Normal | 16 | 10 | 6 | 0 | 0 |
0.00 |
Evaluation of IHC
Immunoreactivity for the Six1 protein was scored
using the staining intensity and positive percentage. Tissue
sections were classed as expressing Six1 if cells showed
immunoreactivity in the nucleus or cytoplasm when observed by an
evaluator that was blinded to the clinical history and outcome. In
total, 10 low-power fields were randomly selected per tissue, and
the cells were counted under a high-power field. The positive
percentage scores were then acquired. Positive percentage scores
were assessed according to the following scale: 0, 0% cells; 1,
0–25% cells; 2, 25–50% cells; and 3, >50% cells. Staining
intensity was then also scored semiquantitatively as follows: 0,
None; 1, mild; 2, moderate; and 3, intense. A total score ranging
between 0 and 9 was then obtained by multiplying the positive
percentage score and intensity score for each research section.
From the total scores, 0, 1–3, 4–6 and 7–9 were recorded as -, +,
++, and +++, respectively. These scores were defined as no or low
expression when the score was <4; positive or high expression
when the score was ≥4. The scores were accepted if two
investigators agreed with the values. Otherwise, the values were
re-estimated until a consensus was reached. The investigators were
in complete agreement in 80% of the cases, which indicated that the
scoring method was highly reproducible.
Statistical analysis
Statistical analysis was performed using the SPSS
statistical software program, version 18.0 (SPSS, Inc., Chicago,
IL, USA). The association between Six1 protein expression and the
clinicopathological data of patients with glioma was estimated
using the χ2 test. The association between survival and
each variable was determined using the Kaplan-Meier method.
Differences between survival rates were analyzed using the log-rank
test and Cox regression analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Six1 expression in 163 glioma
tissues
To identify the Six1 protein expression level, IHC
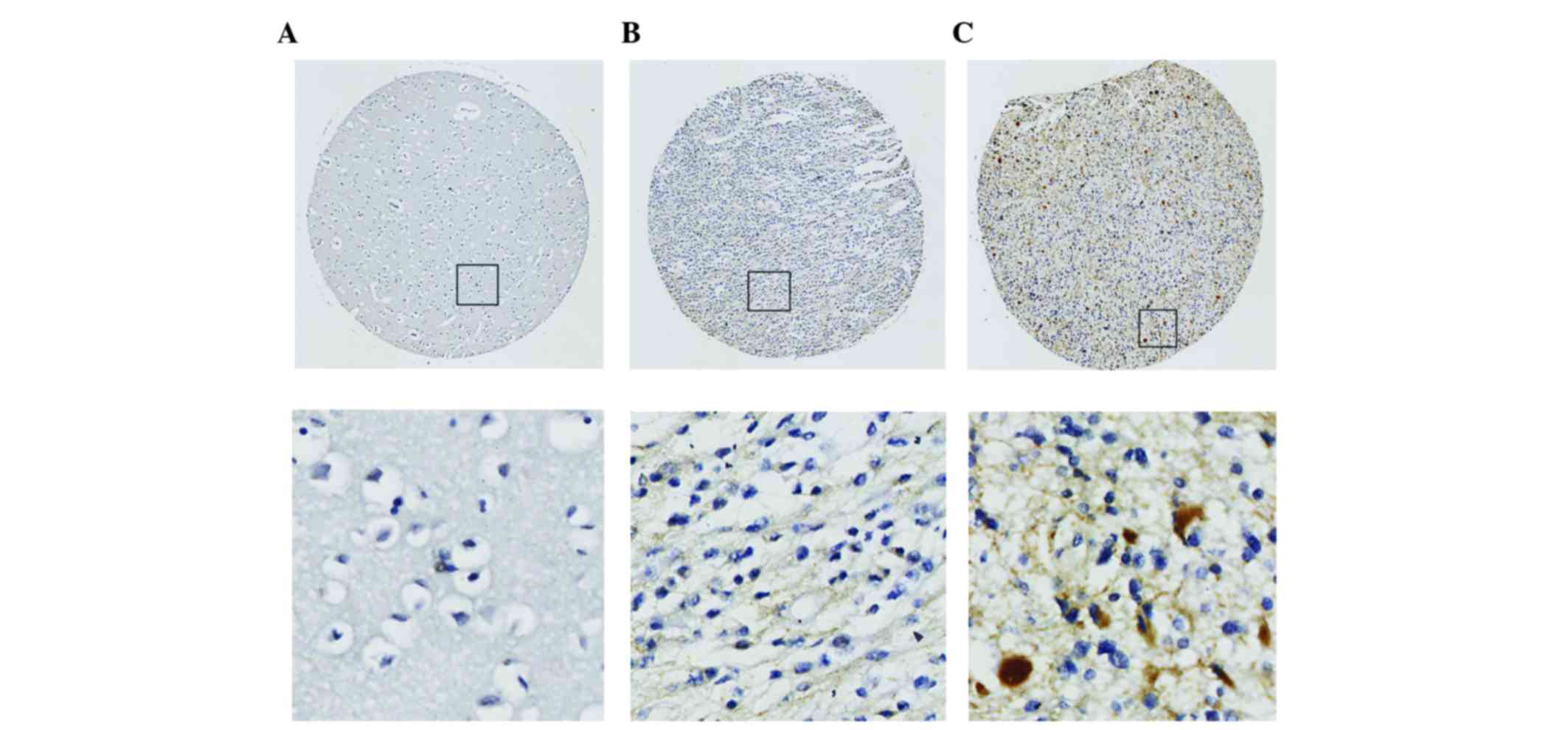
staining was performed. Six1 expression was identified in 49.1% (80
out of 163) of all gliomas. According to the WHO grade, Six1
expression was identified in 34.2% of low-grade (WHO I/II) gliomas
and 80.8% of high-grade (WHO III/IV) gliomas, respectively
(Table I). Overall, the Six1 level in
the high-grade tumors was significantly higher compared with the
level in low-grade tumors (P<0.001; Table I), and the Six1 expression level in
all normal brain tissues was also markedly lower compared with the
level in glioma tissues (P<0.05; Table
I). At the same time, the IHC staining revealed that the Six1
protein was mainly expressed in the cytoplasm (Fig. 1).
Six1 expression and pathological
indicators
As shown in Table II,
Six1 expression was significantly associated with the WHO grade
(P<0.001), indicating that the status of Six1 expression was
upregulated in high-grade glioma patients. No association was
identified between Six1 expression and age. Also, no association
was observed between Six1 expression and gender, indicating that
Six1 expression was not dependent on the gender of the patients. Of
the 4 grades, grade I exhibited the lowest expression level.
However, no significant difference was observed between the
expression of Six1 in grade III and IV gliomas (P=0.084).
Six1 expression was associated with
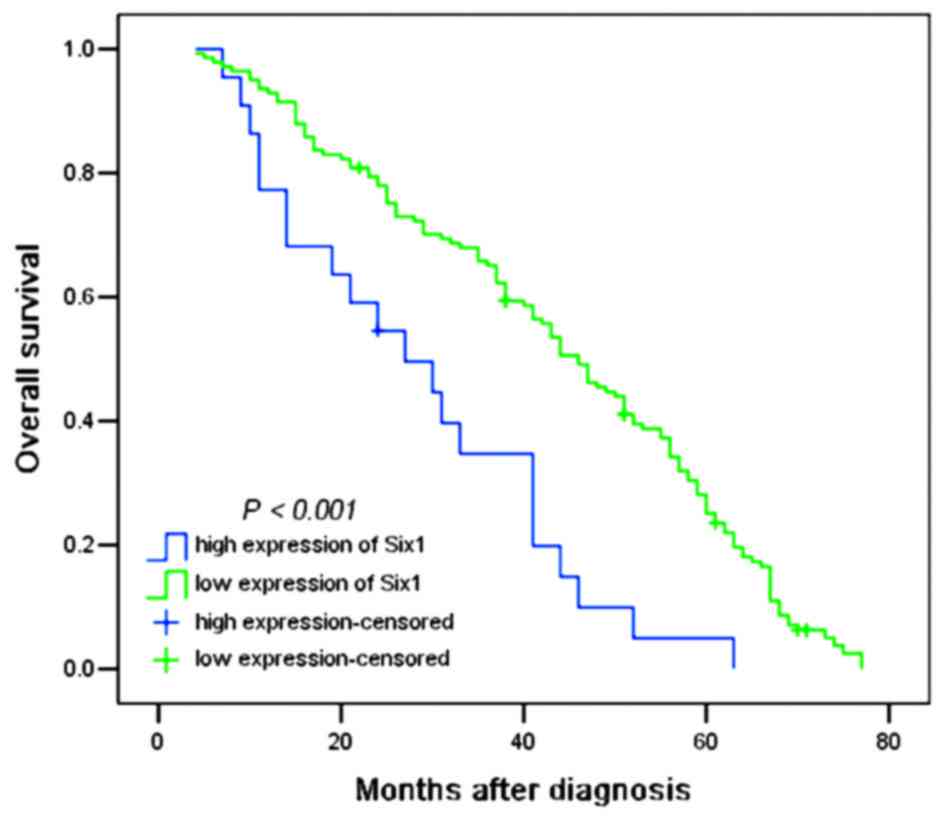
the prognosis of patients
To evaluate the association between Six1 protein
expression and the prognosis of patients, all glioma patients were
allocated to two groups, the low and high Six1 expression groups. A
log-rank test and Kaplan-Meier analysis were performed to assess
the effect of Six1 expression on the patient survival. Out of the
163 patients, the survival data of 153 patients were available,
among which 10 patients were still alive at follow-up and were
censored. The high expression of Six1 in all 141 gliomas exhibited
a significant difference from 21 patients with low expression
(P<0.001; Fig. 2). As shown in
Table III, the median of overall
survival time in all patients was 41.0 months [n=163; 95%
confidence interval (CI), 38.954–47.046]; the median survival time
of patients with low Six1 expression was only 27.0 months (95% CI,
13.892–40.108), whereas the median survival time of those with high
Six1 expression was 46.0 months (95% CI, 40.708–51.292). The
log-rank test revealed that patients with low Six1 expression had a
significantly shorter overall survival time compared with patients
with high Six1 expression (χ2=15.668; P<0.001;
Table III). In addition,
multivariate analysis was also performed to investigate whether
Six1 was an independent prognostic factor for patient survival. As
shown in Table IV, multivariate
analysis identified Six1 expression (P=0.045) and WHO grade
(P<0.001) as independent prognostic factors, instead of age and
gender.
 | Table III.Median survival time of patients with
high and low expression of Six1. |
Table III.
Median survival time of patients with
high and low expression of Six1.
| Six1
expression | Patients, n | Median survival
time, months | 95% CI | χ2 | P-value |
|---|
| Lowa | 141 | 46.00 | 40.708–51.292 | 15.668 | <0.001 |
| High | 22 | 27.00 | 13.892–40.108 |
|
|
| Overall | 163 | 43.00 | 38.954–47.046 |
|
|
 | Table IV.Multivariate analysis of various
prognostic indicators in patients with glioma, performed using the
Cox regression model. |
Table IV.
Multivariate analysis of various
prognostic indicators in patients with glioma, performed using the
Cox regression model.
|
|
| Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | Patients, n | RR | 95% CI | P-value |
|---|
| Median age (≤45
years/>45 years) |
81/82 | 0.728 | 0.522–1.016 | 0.062 |
| Gender
(male/female) |
99/64 | 1.015 | 0.733–1.406 | 0.928 |
| WHO grade
(I+II/III+IV) | 111/52 | 2.695 | 1.838–3.952 | 0.000 |
| Six1 expression
(low/high) | 141/22 | 1.670 | 1.011–2.760 | 0.045 |
Discussion
Homeobox genes encode transcription factors that are
essential for the development of numerous organs and control
processes, such as proliferation, apoptosis, migration, and
invasion (9,21–23). The
Six1 homeoprotein is a member of the Six family of homeodomain
transcription factors and has been found to be upregulated in
multiple cancers, including breast cancer (12,20,24),
rhabdomyosarcomas (18,25,26),
hepatocellular carcinomas (19),
ovarian cancer (13) and Wilms'
tumors (17). In addition, Six1 plays
a role in cellular migration and invasion during embryogenesis
(22,27–30) and in
breast cancer (31,32). Notably, a recent study demonstrated
that messenger RNA profiling of Six1 is dysregulated in
A2B5+ glioma tumor progenitor cells from
A2B5+ glial progenitor cells isolated from normal white
matter (33).
Overexpression of vascular endothelial growth factor
C (VEGF-C) has been detected in numerous cancers (34–37), and
the role of VEGF-C in promoting lymphatic metastasis has been
demonstrated in several VEGF-C overexpression animal models of
mammary carcinoma (38–40). Previous study revealed that Six1 could
coordinate with transforming growth factor-β (TGF-β) to increase
the expression of VEGF-C through two pathways. Firstly, Six1
enhances TGF-β signaling by upregulating TGF-β receptor 1 (TβR1)
expression, which promotes the activation of SMAD family member 2/3
(SMAD2/3) and its binding to the VEGF-C promoter, thus increasing
the expression of VEGF-C. Secondly, Six1 may cooperate with SMAD2/3
to bind to the VEGF-C promoter and modulate VEGF-C expression. In
tumor cells without Six1 expression, the expression of VEGF-C was
not notably affected by TGF-β stimulation, although SMAD2/3 was
phosphorylated and was able to bind to the VEGF-C promoter.
Therefore, Six1 is necessary for TGF-β to induce increased
expression of the VEGF-C gene (41).
In addition, overexpression of Six1 significantly enhances the
activity of the cyclin D1 promoter in pancreatic cancer and
promotes cell cycle progression and proliferation (42). Furthermore, Six1 overexpression is
positively correlated with the disease-free survival and 5-year
overall survival rates of patients with breast cancer (43). However, the expression model and
prognostic value of Six1 in the gliomas were rarely reported.
Therefore, it was hypothesized in the present study that Six1 may
be expressed and play a role in gliomas of different malignancy
grades.
In the present study, the expression of the Six1
protein was detected in glioma tissues of various malignancy
grades, and it was found that the level of Six1 expression in all
glioma tissues was significantly higher than the expression level
in normal brain tissues. Furthermore, Six1 expression was found to
be associated with the WHO grade, but not with age and gender. The
present results indicated that Six1 expression in glioma is
responsible for glioma progress. In order to investigate the effect
of Six1 expression on the prognosis of glioma patients, 163
patients were followed up subsequent to surgery. Six1 was
identified as an independent factor to significantly predict the
overall survival time of glioma patients. Firstly, the log-rank
test revealed that patients with high Six1 expression possess a
significantly shorter median overall survival time of 27 months,
compared with the median of 46 months in the low expression group.
Secondly, Cox regression analysis identified that Six1 may act as
an independent prognostic factor, in addition to the WHO grade.
This indicated that Six1 may be recommended as a useful marker
associated with a worse prognosis in glioma patients.
In conclusion, Six1 is differently expressed in
different grades of glioma and is associated with the WHO grade of
disease, indicating a worse prognosis in patients with glioma. In
addition, the Six1 protein may be suggested as a useful prognostic
biomarker for glioma, including glioblastoma.
Acknowledgements
The authors thank Dr Hongdian Zhang (Department of
Neurosurgery, Affiliated Bayi Brain Hospital, General Hospital of
Beijing Military Region, Beijing, China) for his assistance with
writing the manuscript.
References
|
1
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vecht CJ, Kerkhof M and Duran-Pena A:
Seizure prognosis in brain tumors: New insights and evidence-based
management. Oncologist. 19:751–759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kleihues P and Sobin LH: World Health
Organization classification of tumors. Cancer. 88:2887. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart LA: Chemotherapy in adult
high-grade glioma: A systematic review and meta-analysis of
individual patient data from 12 randomised trials. Lancet.
359:1011–1018. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohgaki H, Dessen P, Jourde B, Horstmann S,
Nishikawa T, Di Patre PL, Burkhard C, Schüler D, Probst-Hensch NM,
Maiorka PC, et al: Genetic pathways to glioblastoma: A
population-based study. Cancer Res. 64:6892–6899. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar JP: The sine oculis homeobox (SIX)
family of transcription factors as regulators of development and
disease. Cell Mol Life Sci. 66:565–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anderson AM, Weasner BM, Weasner BP and
Kumar JP: Dual transcriptional activities of SIX proteins define
their roles in normal and ectopic eye development. Development.
139:991–1000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu PX, Zheng W, Huang L, Maire P, Laclef C
and Silvius D: Six1 is required for the early organogenesis of
mammalian kidney. Development. 130:3085–3094. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Laclef C, Souil E, Demignon J and Maire P:
Thymus, kidney and craniofacial abnormalities in Six 1 deficient
mice. Mech Dev. 120:669–679. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Konishi Y, Ikeda K, Iwakura Y and Kawakami
K: Six1 and Six4 promote survival of sensory neurons during early
trigeminal gangliogenesis. Brain Res. 1116:93–102. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Reichenberger KJ, Coletta RD, Schulte AP,
Varella-Garcia M and Ford HL: Gene amplification is a mechanism of
Six1 overexpression in breast cancer. Cancer Res. 65:2668–2675.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Behbakht K, Qamar L, Aldridge CS, Coletta
RD, Davidson SA, Thorburn A and Ford HL: Six1 overexpression in
ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and
is associated with poor survival. Cancer Res. 67:3036–3042. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Imam JS, Buddavarapu K, Lee-Chang JS,
Ganapathy S, Camosy C, Chen Y and Rao MK: MicroRNA-185 suppresses
tumor growth and progression by targeting the Six1 oncogene in
human cancers. Oncogene. 29:4971–4979. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tan J, Zhang C and Qian J: Expression and
significance of Six1 and Ezrin in cervical cancer tissue. Tumour
Biol. 32:1241–1247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng XH, Liang PH, Guo JX, Zheng YR, Han
J, Yu LL, Zhou YG and Li L: Expression and clinical implications of
homeobox gene Six1 in cervical cancer cell lines and cervical
epithelial tissues. Int J Gynecol Cancer. 20:1587–1592.
2010.PubMed/NCBI
|
|
17
|
Li CM, Guo M, Borczuk A, Powell CA, Wei M,
Thaker HM, Friedman R, Klein U and Tycko B: Gene expression in
Wilms' tumor mimics the earliest committed stage in the metanephric
mesenchymal-epithelial transition. Am J Pathol. 160:2181–2190.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu Y, Khan J, Khanna C, Helman L, Meltzer
PS and Merlino G: Expression profiling identifies the cytoskeletal
organizer ezrin and the developmental homeoprotein Six-1 as key
metastatic regulators. Nature Med. 10:175–181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ng KT, Man K, Sun CK, Lee TK, Poon RT, Lo
CM and Fan ST: Clinicopathological significance of homeoprotein
Six1 in hepatocellular carcinoma. Br J Cancer. 95:1050–1055. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Coletta RD, Christensen K, Reichenberger
KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Müller-Tidow C, Golub
TR, Kawakami K and Ford HL: The Six1 homeoprotein stimulates
tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci USA.
101:6478–6483. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Coletta RD, Christensen KL, Micalizzi DS,
Jedlicka P, Varella-Garcia M and Ford HL: Six1 overexpression in
mammary cells induces genomic instability and is sufficient for
malignant transformation. Cancer Res. 68:2204–2213. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng W, Huang L, Wei ZB, Silvius D, Tang
B and Xu PX: The role of Six1 in mammalian auditory system
development. Development. 130:3989–4000. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ikeda K, Kageyama R, Suzuki Y and Kawakami
K: Six1 is indispensable for production of functional progenitor
cells during olfactory epithelial development. Int J Dev Biol.
54:1453–1464. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ford HL, Kabingu EN, Bump EA, Mutter GL
and Pardee AB: Abrogation of the G2 cell cycle checkpoint
associated with overexpression of HSIX1: A possible mechanism of
breast carcinogenesis. Proc Natl Acad Sci USA. 95:12608–12613.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khan J, Bittner ML, Saal LH, Teichmann U,
Azorsa DO, Gooden GC, Pavan WJ, Trent JM and Meltzer PS: cDNA
microarrays detect activation of a myogenic transcription program
by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA.
96:13264–13269. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu Y, Davicioni E, Triche TJ and Merlino
G: The homeoprotein six1 transcriptionally activates multiple
protumorigenic genes but requires ezrin to promote metastasis.
Cancer Res. 66:1982–1989. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ozaki H, Nakamura K, Funahashi J, Ikeda K,
Yamada G, Tokano H, Okamura HO, Kitamura K, Muto S, Kotaki H, et
al: Six1 controls patterning of the mouse otic vesicle.
Development. 131:551–562. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Oghi KA, Zhang J, Krones A, Bush KT,
Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW and Rosenfeld MG:
Eya protein phosphatase activity regulates Six1-Dach-Eya
transcriptional effects in mammalian organogenesis. Nature.
426:247–254. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Grifone R, Demignon J, Houbron C, Souil E,
Niro C, Seller MJ, Hamard G and Maire P: Six1 and Six4
homeoproteins are required for Pax3 and Mrf expression during
myogenesis in the mouse embryo. Development. 132:2235–2249. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ikeda K, Ookawara S, Sato S, Ando Z,
Kageyama R and Kawakami K: Six1 is essential for early neurogenesis
in the development of olfactory epithelium. Dev Biol. 311:53–68.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Micalizzi DS, Christensen KL, Jedlicka P,
Coletta RD, Barón AE, Harrell JC, Horwitz KB, Billheimer D,
Heichman KA, Welm AL, et al: The Six1 homeoprotein induces human
mammary carcinoma cells to undergo epithelial-mesenchymal
transition and metastasis in mice through increasing TGF-beta
signaling. J Clin Invest. 119:2678–2690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Micalizzi DS, Wang CA, Farabaugh SM,
Schiemann WP and Ford HL: Homeoprotein Six1 increases TGF-beta type
I receptor and converts TGF-beta signaling from suppressive to
supportive for tumor growth. Cancer Res. 70:10371–10380. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Auvergne RM, Sim FJ, Wang S,
Chandler-Militello D, Burch J, Al Fanek Y, Davis D, Benraiss A,
Walter K, Achanta P, et al: Transcriptional differences between
normal and glioma-derived glial progenitor cells identify a core
set of dysregulated genes. Cell Rep. 3:2127–2141. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang J, Wu HF, Qian LX, Zhang W, Hua LX,
Yu ML, Wang Z, Xu ZQ, Sui YG and Wang XR: Increased expressions of
vascular endothelial growth factor (VEGF), VEGF-C and VEGF
receptor-3 in prostate cancer tissue are associated with tumor
progression. Asian J Androl. 8:169–175. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ueda M, Terai Y, Yamashita Y, Kumagai K,
Ueki K, Yamaguchi H, Akise D, Hung YC and Ueki M: Correlation
between vascular endothelial growth factor-C expression and
invasion phenotype in cervical carcinomas. Int J Cancer.
98:335–343. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
O-charoenrat P, Rhys-Evans P and Eccles
SA: Expression of vascular endothelial growth factor family members
in head and neck squamous cell carcinoma correlates with lymph node
metastasis. Cancer. 92:556–568. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kinoshita J, Kitamura K, Kabashima A,
Saeki H, Tanaka S and Sugimachi K: Clinical significance of
vascular endothelial growth factor-C (VEGF-C) in breast cancer.
Breast Cancer Res Treat. 66:159–164. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Karpanen T, Egeblad M, Karkkainen MJ, Kubo
H, Ylä-Herttuala S, Jäättelä M and Alitalo K: Vascular endothelial
growth factor C promotes tumor lymphangiogenesis and intralymphatic
tumor growth. Cancer Res. 61:1786–1790. 2001.PubMed/NCBI
|
|
39
|
Mattila MM, Ruohola JK, Karpanen T,
Jackson DG, Alitalo K and Härkönen PL: VEGF-C induced
lymphangiogenesis is associated with lymph node metastasis in
orthotopic MCF-7 tumors. Int J Cancer. 98:946–951. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Skobe M, Hawighorst T, Jackson DG, Prevo
R, Janes L, Velasco P, Riccardi L, Alitalo K, Claffey K and Detmar
M: Induction of tumor lymphangiogenesis by VEGF-C promotes breast
cancer metastasis. Nat Med. 7:192–198. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu D, Li L, Zhang XX, Wan DY, Xi BX, Hu
Z, Ding WC, Zhu D, Wang XL, Wang W, et al: SIX1 promotes tumor
lymphangiogenesis by coordinating TGFβ signals that increase
expression of VEGF-C. Cancer Res. 74:5597–5607. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang
L, Li X, Li L, Ma W, Wu J and Zhang M: Six1 promotes proliferation
of pancreatic cancer cells via upregulation of cyclin D1
expression. PloS one. 8:e592032013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin H, Cui M, Kong J, Cui X, Lin Z, Wu Q
and Liu S: Sineoculis homeobox homolog 1 protein is associated with
breast cancer progression and survival outcome. Exp Mol Pathol.
97:247–252. 2014. View Article : Google Scholar : PubMed/NCBI
|