Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer and the third leading cause of cancer deaths
worldwide with a <7% five-year survival rate (1–3). Globally,
infection by the hepatitis B virus (HBV) is the most prevalent
cause of HCC. The prognosis of HCC can be influenced by several
clinicopathological factors, such as liver function, number of
nodules, tumor size, vascular invasion and differentiation grade
(4–6).
Among these, tumor differentiation grade is the most significant
factor in deciding on the type of therapy that HCC patients should
receive. The most widely used method to assess differentiation
grade are the Edmondson-Steiner criteria (7). However, this is currently achieved using
liver biopsy, which may increase the risk of HCC further
developing, for example, exposing patients to the risk of tumor
cell-seeding (8). In addition, needle
biopsies have a significant false-negative rate for the diagnosis
of HCC due to sampling errors (9).
Furthermore, determination of the Edmondson grades is likely to be
affected by subjective factors and therefore, additional studies
that focus on the exploration of non-invasive prediction of
differentiation are required.
Recent advances in analytical chemistry have
resulted in metabolomics becoming an important way of understanding
disease mechanisms and identifying candidate biomarkers.
Metabolomics is defined as the comprehensive quantitative and
qualitative analysis of all metabolites in cells, tissues, or
biofluids in response to biological interventions or environmental
factors (10,11). An increasing number of studies focus
on the pathogenesis and biomarkers of HCC using metabolomics
methodology. Thus metabolomics may provide a way to select
characteristic metabolites that can distinguish HCC patients with
diverse differentiation grades without having to carry out a
biopsy.
In the current study, an ultra performance liquid
chromatography and linear trap quadrupole (UPLC-LTQ)-Orbitrap XL
mass spectrometry (MS) analytical platform was used to analyze the
serum metabolic profiling of HBV-related HCC patients with diverse
differentiation grades. Following selection and identification of
the metabolites, their clinical value was assessed by receiver
operator characteristic curve (ROC) analysis and area under the
curve (AUC) analysis.
Materials and methods
Chemicals and instruments
All solvents used in the present study were high
performance liquid chromatography (HPLC) grade without
modification. Formic acid and acetonitrile were obtained from Merck
(Merck Millipore, Darmstadt, Germany). Distilled water was produced
using a Milli-Q Reagent Water System (EMD Millipore, Billerica, MA,
United States). Standard preparations of Lysophosphatidylcholine
(LysoPC) (16:0) and LysoPC (18:0) were purchased from Sigma-Aldrich
(St. Louis, MO, USA). Calibration standards [caffeine, Ultramark
1621 and methionine-arginine-phenylalanine-alanine (MRFA)] were
provided by Thermo Fisher Scientific Inc. (Waltham MA, United
States). UPLC was performed using an Accela system (Thermo Fisher
Scientific Inc. Waltham, MA, United States). MS was performed with
a LTQ Orbitrap XL hybrid mass spectrometer (Thermo Fisher
Scientific Inc.).
Enrolled population and sample
collection
A total of 58 HBV-related HCC patients were included
in the current study. All samples were obtained between October
2013 and March 2015 from inpatients and outpatients at the Tianjin
Third Central Hospital, Tianjin, China. HCC was diagnosed in
accordance with clinical practice guidelines proposed in 2012
(12), and the diagnostic results
were confirmed by histopathological examination. The patients were
divided into three groups according to the Edmondson-Steiner
criteria: Group A included 21 patients with high-grade HCC
(Edmondson-Steiner I), group B included 23 patients with
middle-grade HCC (Edmondson-Steiner II), and group C included 14
patients with low-grade HCC (Edmondson-Steiner III/IV). The
pathological images of HCC tissue are presented in Fig. 1. A further 20 patients were diagnosed
with HBV-related liver cirrhosis (LC), and were all classified as
Child-Pugh A grade (13). These
patients were put into a different group, called Group LC. All of
the patients within the current study were confirmed to have no
secondary liver cancer, other systemic tumors, diabetes or other
metabolic diseases. A control group was also included, consisting
of 19 healthy volunteers, all were confirmed to have normal liver
function, no viral hepatitis, no alcohol or nonalcohol fatty liver,
no metabolic disease or other complications. The demographic and
clinical characteristics of all participants are presented in
Table I.
 | Table I.Clinical parameters for HCC, LC, and
control groups. |
Table I.
Clinical parameters for HCC, LC, and
control groups.
|
| HCC group (n=58) |
|
|
|---|
|
|
|
|
|
|---|
| Parameters | Group A (n=21) | Group B (n=23) | Group C (n=14) | Group LC (n=20) | Control group
(n=19) |
|---|
| Male/female | 14/7 | 17/6 | 11/3 | 14/6 | 12/7 |
| Age | 56.4±5.4 | 56.7±6.2 | 55.3±5.8 | 56.3±5.3 | 57.0±6.1 |
| ALB (g/l) | 40.30
(37.90–47.30) | 40.80
(39.55–44.85) | 40.55
(39.17–43.57) | 41.23
(37.54–42.57) | 47.70
(45.4–49.40) |
| ALT (U/l) | 33.00
(19.00–60.13) | 39.00
(22.00–79.50) | 47.00
(26.50–78.50) | 29 (19.00–40.00) | 18.00
(15.00–21.00) |
| AST (U/l) | 34.00
(27.00–57.50) | 34.00
(22.00–71.50) | 39.00
(16.75–75.84) | 27.5
(18.00–43.25) | 20.00
(17.00–24.00) |
| GGT (U/l) | 56.00
(32.00–78.00) | 71.00
(45.50–160.00) | 103.50
(58.75–147.50) | 35 (19.00–53.00) | 16.00
(13.00–26.00) |
| TBA (µmol/l) | 8.70
(4.10–11.87) | 8.80
(4.15–22.14) | 9.00
(6.55–16.02) | 0.86 (0.50–1.87) | 1.90
(1.200–2.400) |
| AFP (ng/ml) | 145.29
(3.96–897.29) | 156.45
(2.34–1103.23) | 152.97
(12.14–1210.24) | 3.19 (1.21–9.85) | 2.71 (2.11–3.38) |
None of the participants were on any medical
treatment for the 4 weeks prior to sample collection. Fasting blood
samples were collected and centrifuged at 2,000 × g for 30
min. The sera were separated, aliquoted and stored at −80°C.
The study protocol was approved by the Hospital
Ethics Committee of Tianjin Third Central Hospital and adhered to
the tenets of the Declaration of Helsinki. All participants
voluntarily joined this study and provided their informed consent
prior to commencing any study procedures.
Sample preparation
Prior to UPLC/MS analysis, serum samples were thawed
at room temperature and an aliquot of 100 ml of serum sample was
mixed with 300 ml methanol. The mixture was vibrated for 30s and
then left to stand for 45 min at room temperature. The mixture was
then centrifuged at 4°C at 10,000 × g for 30 min. The
supernatant was filtered using a 0.22 µm membrane (Merck Millipore,
Darmstadt, Germany).
Sample analysis
Chromatography was performed using an Accela system
(Thermo Fisher Scientific Inc.) equipped with a binary solvent
delivery manager and a sample manager. The analytical column was a
Thermo Hypersil GOLD (2.1 mm xID50 mm, 1.9 µm) C18
reverse phase column. The injected volume was 10 µl and the flow
rate was maintained at 200 µl/min. The temperatures of the sample
manager and column oven were set at 4°C and 20°C, respectively. For
UPLC analysis, the mobile phase consisted of 0.1% formic acid
aqueous solution (phase A) and 0.1% formic acid acetonitrile
solution (phase B) (Merck Millipore). Chromatographic separation
was performed within 21 min per sample: i) 95% phase A and 5% phase
B were held for 2.5 min initially; ii) Phase B was gradually
escalated to 95% in the following 7 min; iii) Phase B was
maintained at 95% for 3 min and then gradually reduced to 5% in the
following 6 min; iv) 5% phase B was held for 2.5 min to balance the
analytical column.
MS was performed in positive mode. The following
parameters were employed: Ion spray voltage, 4.5 kV; capillary
voltage, 30 V; cone voltage, 150 V; desolvation temperature, 350°C;
sheath gas flow rate of 30 arb and assistant gas flow rate of 5 arb
(99.999% nitrogen). Data were collected in centroid mode and the
mass-to-charge ratio (m/z) range was set at 50–1000. MS resolution
was 100,000 full width at half maximum (FWHM) and calibration
standards were used, including caffeine, Ultramark 1621 and MRFA
(Thermo Fisher Scientific Inc.). MS/MS analysis was performed by
using collision-induced dissociation at 35% normalization collision
energy and the collision gas was 99.999% helium.
Statistical analysis
MZmine 2.0 software (14) was used for peak detection, alignment
and normalization. The filter conditions were: Each chromatography
peak signal-to-noise ratio >30, the retention time tolerance at
± 0.1 min and the m/z tolerance at ±0.01 Da. SIMCA-P+ ver. 12.0.1.0
software (Umetrics, Malmö, Sweden) was used to establish the
principal component analysis (PCA) and orthogonal partial least
squares discriminant analysis (OPLS-DA) model of all the samples
and the result was checked by cross validation described previously
(15,16). Preliminary selection of characteristic
metabolites was accomplished using the corresponding variable
influence on projection (VIP) value, confidence interval and
coefficient plot generated by the OPLS-DA model. SPSS ver. 17.0
software (SPSS Inc., Chicago, IL United States) was used to
evaluate the statistical significance of differences of the
variances among diverse groups. The ROC curves were generated and
the corresponding AUC was calculated. P<0.05 was considered to
indicate a statistically significant difference.
Identification of the characteristic
metabolites
Some characteristic ions were identified by
comparing the secondary mass spectrum and retention index with
authentic reference standards. Due to the high resolution of the
Orbitrap XL MS (100,000 FWHM), other characteristic metabolites
were identified by checking the accurate m/z value against the
Human Metabolome DataBase (http://hmdb.ca).
Matching substances that had m/z deviations <0.01 were
considered for further identification when the ionization modes
were the same as those used in the HMDB database. After MS/MS
scanning, the secondary mass spectrometer (MS2) spectra of
characteristic ions were obtained and compared with theoretical
fragments from preliminary results. This established that the MS2
m/z deviation was <0.2, the top three peaks matched and that
there was ≥80% match between the preliminary and secondary mass
spectra. The theoretical fragments were derived using Mass Frontier
6.0 software (Thermo Fisher Scientific Inc.).
Results
Metabolic profile
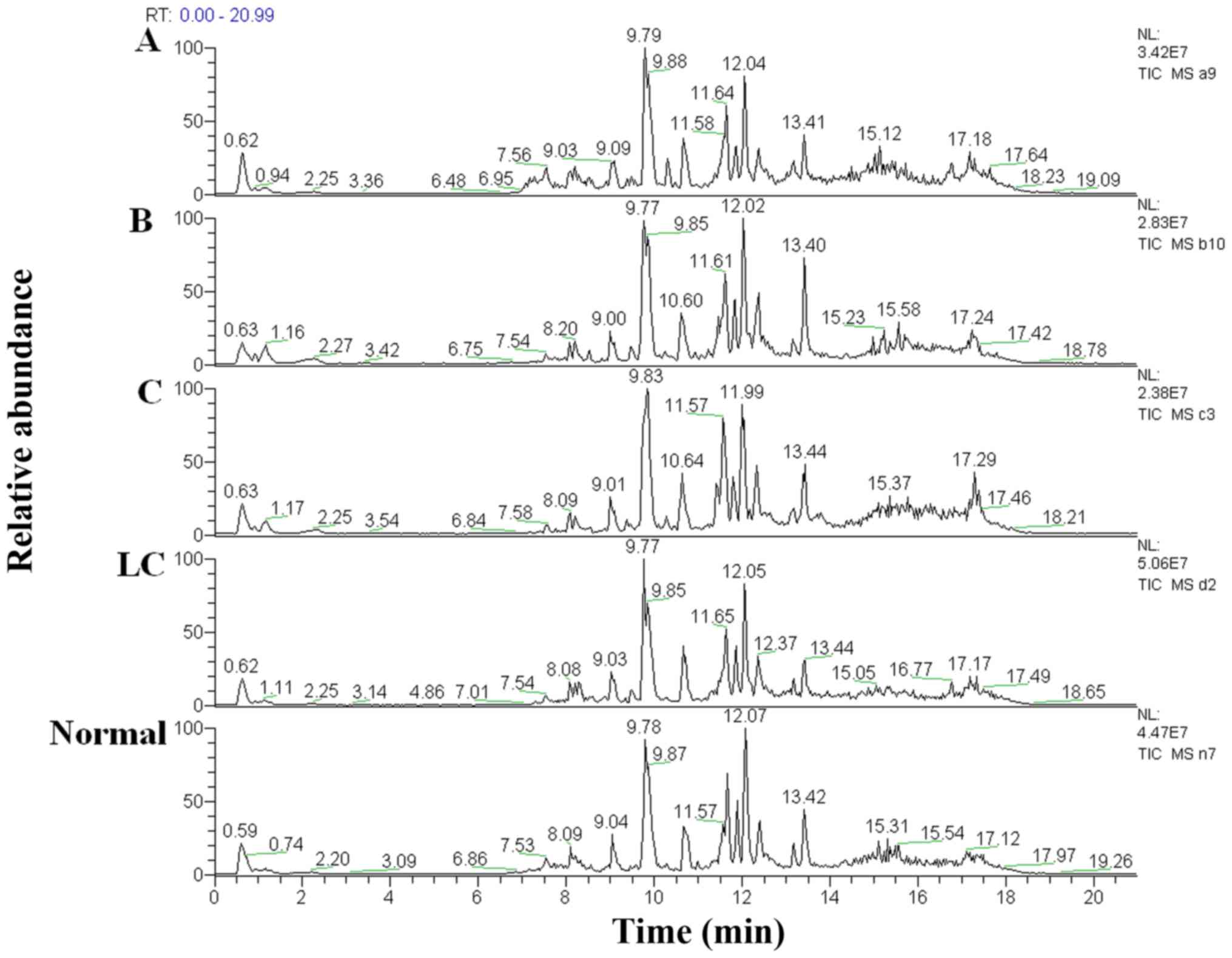
The total ion chromatograms of a single sample from
each patient group acquired by the UPLC-MS platform are presented
in Fig. 2. Using the MZmine ver. 2.0
software, this pre-treatment sought out 950 integral peaks
following extraction ion chromatography detection in all samples.
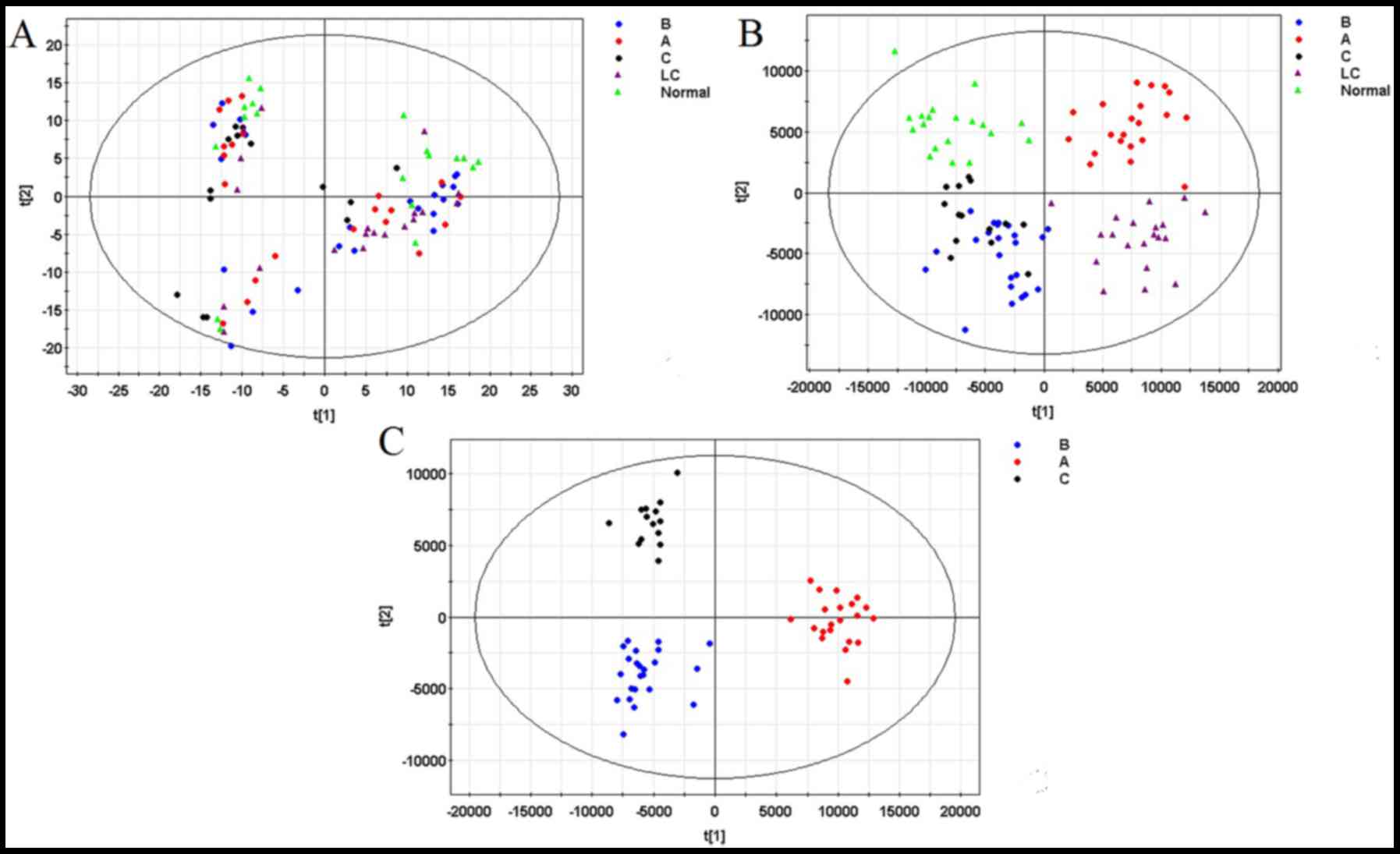
Following pre-treatment, a PCA model was established (R2X=44.7%,
Q2=19.4%), as well as an OPLS-DA model (R2X=74.5%, R2Y=82.5%,
Q2=49.6%), using data from all groups. No significant outliers were
identified in the PCA model, indicating the pretreatment
consistency was satisfactory and the analysis-detecting system was
stable. However the PCA model did not show significant clustering
tendency (Fig. 3A). In contrast, the
OPLS-DA model (Fig. 3B), which
constituted with the first predictive principal component and
second predictive principal component, revealed a clustering
tendency of data from each group.
Selection and identification of
characteristic metabolites
For the purpose of exploring endogenous
characteristic metabolites relating to differentiation grade, the
characteristic metabolites affecting the clustering tendency of the
OPLS-DA model within groups A, B and C were selected following two
steps: i) A new OPLS-DA model was established (R2X=66.2%,
R2Y=91.1%, Q2=64.7%) based on the three HCC groups (Fig. 3C); ii) Ions with a variable importance
value (VIP) >1 were selected, and the ions which included zero
in the confidence interval in the VIP diagram or the coefficient
plot were excluded as previously described (17). Following these steps, 14 ions were
selected as candidates for identification.
According to the process mentioned previously
(18), 5 of the 14 selected ions were
identified. The final results and statistical differences among the
three HCC groups are presented in Table
II.
 | Table II.Metabolite identification. |
Table II.
Metabolite identification.
|
|
| Contentc |
|---|
|
|
|
|
|---|
| Metabolite | Adductb | B/A | C/B | C/A |
|---|
| LysoPC
(16:0)a |
[M+H]+ | Down (P=0.01) |
| Down (P=0.001) |
| Oleamide |
[M+Na]+ | Down (P=0.001) | Down (P=0.001) | Down (P=0.001) |
| MG
(0:0/15:0/0:0) |
[M+NH4]+ |
| Down (P=0.003) | Down (P=0.001) |
| LysoPC (18:0) |
[M+Na]+ |
| Down (P=0.001) | Down (P=0.001) |
| LysoPC
[22:5(7Z,10Z,13Z,16Z,19Z)] |
[M+H]+ |
|
| Down*
(P=0.003) |
ROC analysis
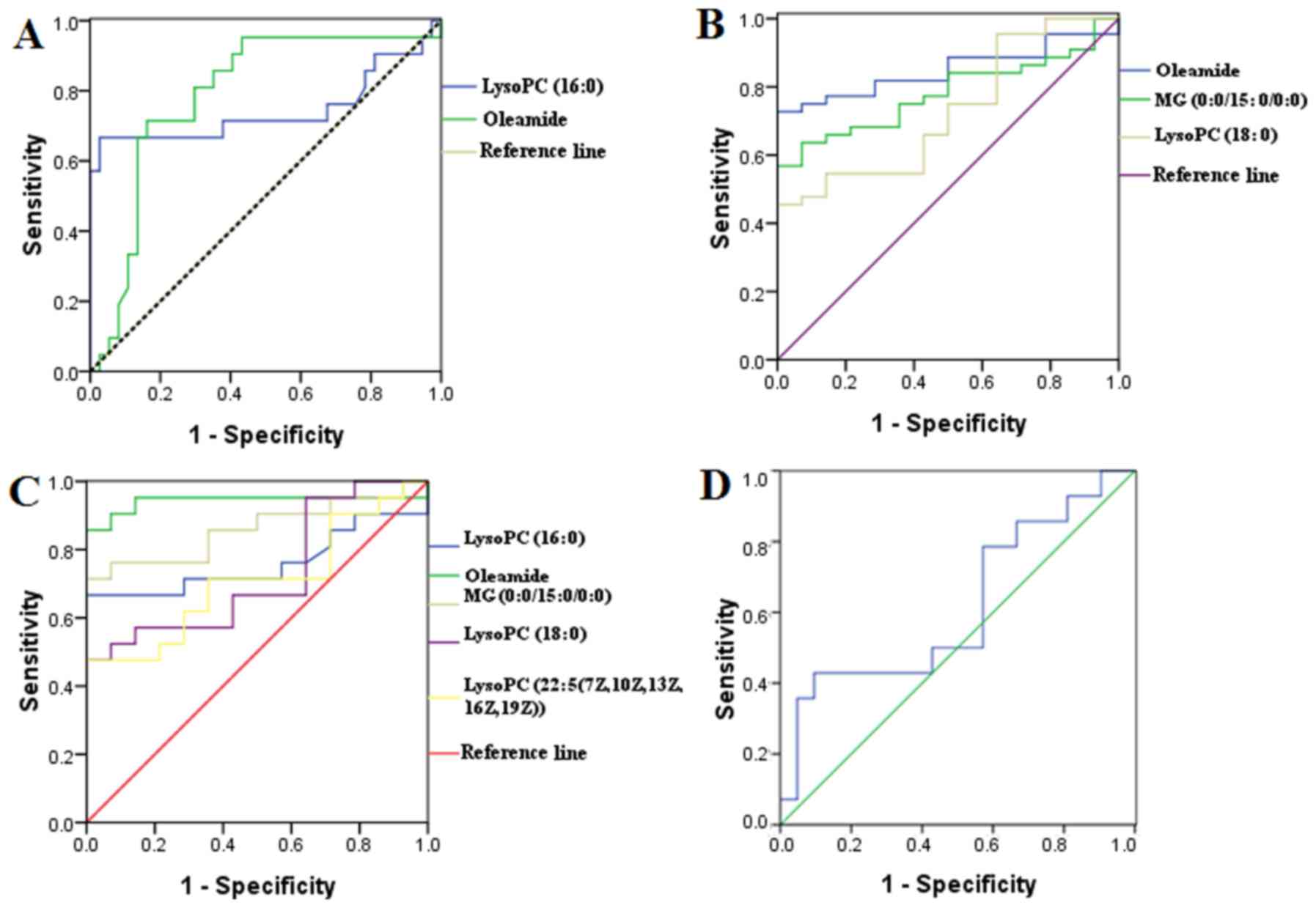
The ROC curves were generated from the identified
metabolites and alpha-fetoprotein (AFP; Fig. 4) to uncover the specificity of the
previously identified metabolites in distinguishing HCC patients
with diverse differentiation grades, and therefore their clinical
value. The AUC of LysoPC (16:0) and Oleamide for distinguishing
Group A from Groups B and C were 0.743 and 0.788 respectively.
Moreover, the AUC of Oleamide, MG (0:0/15:0/0:0) and LysoPC (18:0)
for distinguishing Group C from Groups A and B were 0.849, 0.781
and 0.727 respectively. In addition the AUC of LysoPC (16:0),
Oleamide, monoglyceride (MG) (0:0/15:0/0:0), LysoPC (18:0) and
LysoPC [22:5(7Z,10Z,13Z,16Z,19Z)] for distinguishing Group A from
Group C were 0.760, 0.942, 0.861, 0.728 and 0.707 respectively.
However, the AUC of AFP for distinguishing Group A from Group C was
0.616, with no significant difference.
Discussion
Identifying the correct differentiation grade of HCC
is an important basis for the selection of an effective treatment
that provides patients with the best long-term prognosis. DuBay
et al (19) have previously
reported that surgical resection of poorly differentiated HCC could
increase patient mortality rates by 25 times. As reported
previously, low-grade differentiation is an independent prognostic
factor for HCC patients (20).
Therefore, there is a clinical imperative to establish an effective
strategy for identifying HCC patients with diverse differentiation
grades. Histological methods such as biopsies are invasive and not
generally accepted by patients. Imaging parameters, including
contrast-enhanced ultrasound washout rate and apparent diffusion
coefficient value, are related to HCC differentiation grades
(21,22), but remain unsatisfactory.
Metabolomics, a top-down systems biology approach,
is often used to explore disease mechanisms and search for
candidate diagnostic biomarkers. Therefore, the present study
focused on the metabolic profile of HCC patients with diverse
differentiation grades, with the aim of selecting characteristic
metabolites to accurately identify each differentiation grade.
The five metabolites identified are lipid
metabolites involved in inflammation and signal transduction, and
it is well known that the liver is important in lipid metabolism.
Unfortunately, the results could not explain why the amounts of
metabolites differed among the three groups. To understand how
these metabolites work in patients with different types of HCC,
further studies are required.
LysoPC is generated by the action of phospholipase
A2 on membrane phosphatidylcholine, the most abundant cellular
phospholipid (23). LysoPC has been
reported to be decreased in patients with HCC (24). Compared with the control group, LysoPC
[22:5(7Z,10Z,13Z,16Z,19Z)] was decreased in the HCC patients group
(not shown). All three types of LysoPC demonstrated an ability to
distinguish HCC patients with diverse differentiation grades.
Decreased LysoPC levels may therefore indicate poorly
differentiated HCC.
Oleamide is the prototype long chain primary fatty
acid amide lipid messenger, first identified in human serum in 1989
(25). It is known to mediate the
drive to sleep, has profound effects on thermoregulation, and acts
as an analgesic in several models of experimental pain (25). The present study demonstrated a
reduction in Oleamide levels when the differentiation degree
decreased. Although its association with differentiation degree
remains unclear, the ability of Oleamide to distinguish HCC
patients with diverse differentiation merits further study.
In conclusion, the successful establishment of a
metabolic profiling model revealed that utilizing metabolomics is a
promising approach to find characteristic metabolites in serum that
may be used to distinguish HCC patients with diverse
differentiation grades. The 5 characteristic metabolites
identified, Lysophosphatidylcholine (16:0), Oleamide, Monoglyceride
(0:0/15:0/0:0), Lysophosphatidylcholine (18:0) and
Lysophosphatidylcholine [22:5(7Z,10Z,13Z,16Z,19Z)], have this
ability. Future research should focus on these characteristic
metabolites to confirm their clinical value. In addition, research
into the pathways related to these metabolites would provide
reliable evidence supporting their use clinically.
Acknowledgements
The present study was supported by the Tianjin
Research Program of Application Foundation and Advanced Technology
(grant no. 13JCYBJC22100), Tianjin Research Program of Application
Foundation and Advanced Technology (grant no. 16JCQNJC11600) and
the General Item of Science and Technology Fund of Tianjin
Municipal Bureau of Health (grant no. 2014KY01). The authors also
thank Y. Wu (Clinical Laboratory Department Third Central Clinical
College, Tianjin Medical University, Tianjin, China) for
support.
References
|
1
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel M, Shariff MI, Ladep NG,
Thillainayagam AV, Thomas HC, Khan SA and Taylor-Robinson SD:
Hepatocellular carcinoma: Diagnostics and screening. J Eval Clin
Pract. 18:335–342. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Y, Hong Z, Tan G, Dong X, Yang G, Zhao
L, Chen X, Zhu Z, Lou Z, Qian B, et al: NMR and LC/MS-based global
metabolomics to identify serum biomarkers differentiating
hepatocellular carcinoma from liver cirrhosis. Int J Cancer.
135:658–668. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiang L, Huikai L, Butt K, Wang PP and Hao
X: Factors associated with disease survival after surgical
resection in Chinese patients with hepatocellular carcinoma. World
J Surg. 30:439–445. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shah SA, Greig PD, Gallinger S, Cattral
MS, Dixon E, Kim RD, Taylor BR, Grant DR and Vollmer CM: Factors
associated with early recurrence after resection for hepatocellular
carcinoma and outcomes. J Am Coll Surg. 202:275–283. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martins A, Cortez-Pinto H, Marques-Vidal
P, Mendes N, Silva S, Fatela N, Glória H, Marinho R, Távora I,
Ramalho F and de Moura MC: Treatment and prognostic factors in
patients with hepatocellular carcinoma. Liver Int. 26:680–687.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Edmondson HA and Steiner PE: Primary
carcinoma of the liver: A study of 100 cases among 48,900
necropsies. Cancer. 7:462–503. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stigliano R, Marelli L, Yu D, Davies N,
Patch D and Burroughs AK: Seeding following percutaneous diagnostic
and therapeutic approaches for hepatocellular carcinoma. What is
the risk and the outcome? Seeding risk for percutaneous approach of
HCC. Cancer Treat Rev. 33:437–447. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pawlik TM, Gleisner AL, Anders RA,
Assumpcao L, Maley W and Choti MA: Preoperative assessment of
hepatocellular carcinoma tumor grade using needle biopsy:
Implications for transplant eligibility. Ann Surg. 245:435–442.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nicholson JK, Lindon JC and Holmes E:
‘Metabonomics’: Understanding the metabolic responses of living
systems to pathophysiological stimuli via multivariate statistical
analysis of biological NMR spectroscopic data. Xenobiotica.
29:1181–1189. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fiehn O: Metabolomics-the link between
genotypes and phenotypes. Plant Mol Biol. 48:155–171. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
European Association of the Study of the
Liver, . 2011 European Association of the Study of the Liver
hepatitis C virus clinical practice guidelines. Liver Int.
32:(Suppl 1). 2–8. 2012. View Article : Google Scholar
|
|
13
|
de Bruijne J, Buster EH, Gelderblom HC,
Brouwer JT, de Knegt RJ, van Erpecum KJ, Schalm SW, Bakker CM,
Zaaijer HL, Janssen HL, et al: Treatment of chronic hepatitis C
virus infection-Dutch national guidelines. Neth J Med. 66:311–322.
2008.PubMed/NCBI
|
|
14
|
Pluskal T, Castillo S, Villar-Briones A
and Oresic M: MZmine 2: Modular framework for processing,
visualizing, and analyzing mass spectrometry-based molecular
profile data. BMC Bioinformatics. 11:3952010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Trygg J, Holmes E and Lundstedt T:
Chemometrics in metabonomics. J Proteome Res. 6:469–479. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Eriksson L, Johansson E, Lindgren F,
Sjostrom M and Wold S: Megavariate analysis of hierarchical QSAR
data. J Comput Aided Mol Des. 16:711–726. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yin P, Wan D, Zhao C, Chen J, Zhao X, Wang
W, Lu X, Yang S, Gu J and Xu G: A metabonomic study of hepatitis
B-induced liver cirrhosis and hepatocellular carcinoma by using
RP-LC and HILIC coupled with mass spectrometry. Mol Biosyst.
5:868–876. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu SY, Zhang RL, Kang H, Fan ZJ and Du Z:
Human liver tissue metabolic profiling research on hepatitis B
virus-related hepatocellular carcinoma. World J Gastroenterol.
19:3423–3432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
DuBay D, Sandroussi C, Sandhu L, Cleary S,
Guba M, Cattral MS, McGilvray I, Ghanekar A, Selzner M, Greig PD
and Grant DR: Liver transplantation for advanced hepatocellular
carcinoma using poor tumor differentiation on biopsy as an
exclusion criterion. Ann Surg. 253:166–172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yaprak O, Akyildiz M, Dayangac M, Demirbas
BT, Guler N, Dogusoy GB, Yuzer Y and Tokat Y: AFP level and
histologic differentiation predict the survival of patients with
liver transplantation for hepatocellular carcinoma. Hepatobiliary
Pancreat Dis Int. 11:256–261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang T, Wang J, Luo Z, Zhu K, Chen J and
Shan H: Study of apparent diffusion coefficient value and
histopathological differentiation of hepatocellular carcinoma.
Zhonghua Yi Xue Za Zhi. 95:187–191. 2015.(In Chinese). PubMed/NCBI
|
|
22
|
Feng Y, Qin XC, Luo Y, Li YZ and Zhou X:
Efficacy of contrast-enhanced ultrasound washout rate in predicting
hepatocellular carcinoma differentiation. Ultrasound Med Biol.
41:1553–1560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ryborg AK, Johansen C, Iversen L and
Kragballe K: Lysophosphatidylcholine induces keratinocyte
differentiation and upregulation of AP-1- and NF-kappaB DNA-binding
activity. Acta Derm Venereol. 84:433–438. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou L, Ding L, Yin P, Lu X, Wang X, Niu
J, Gao P and Xu G: Serum metabolic profiling study of
hepatocellular carcinoma infected with hepatitis B or hepatitis C
virus by using liquid chromatography-mass spectrometry. J Proteome
Res. 11:5433–5442. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mueller GP and Driscoll WJ: Biosynthesis
of oleamide. Vitam Horm. 81:55–78. 2009. View Article : Google Scholar : PubMed/NCBI
|