Introduction
Esophageal cancer is among the most prevalent
cancers in China, ranking sixth in terms of incidence and fourth in
terms of mortality, according to the 2011 Annual Report on the
Status of Cancer in China (1). The
highest morbidity rate of esophageal cancer in China reached 130
per 100,000 people in 2003 (2).
Esophageal cancer can be further divided into two main pathological
subtypes: Esophageal squamous cell carcinoma (ESCC) and esophageal
adenocarcinoma (EAC), with the former being the predominant type
and accounting for ~90% of esophageal cancer cases worldwide
(3). Even though there have been
significant advances in therapeutic approaches for ESCC, the
prognosis of ESCC remains poor (the 5-year survival rate is
<10%) (4). Therefore, it is
essential to explore the pathogenesis of ESCC and to identify novel
biomarkers for risk assessment, early detection and prognosis
prediction.
Many genetic and epigenetic alterations have been
implicated in the pathogenesis of ESCC. Among these alterations,
DNA methylation-induced gene silencing of tumor suppressor genes
plays significant roles in ESCC initiation, progression and
metastasis; thus serving as important ESCC biomarkers (5). Although expression analysis alone is
frequently used as the main approach to identify cancer biomarkers,
Cheng et al recently proposed that integrating expression
and epigenetic alteration analyses would provide a higher
likelihood of identifying ESCC biomarkers (6).
Raf kinase inhibitory protein (RKIP) is a highly
conserved, ubiquitously expressed and small cytoplasmic protein
with various biological and pathological activities (7). RKIP inhibits Raf-1-mediated
phosphorylation and activation of mitogen-activated protein kinase
(MAPK) kinase (MEK)-1, as well as the subsequent MAPK and
extracellular signal-regulated kinase activities (8). RKIP also negatively regulates nuclear
factor (NF)-κB signaling and the signaling downstream of G
protein-coupled receptor kinase (9,10). During
cancer development, RKIP is characterized as a tumor suppressor
gene because of its maintenance of chromosome stability, inhibition
of cellular proliferation, promotion of cell differentiation and
dynamic balancing of oncogene activities (11). Consistently, previous studies have
reported RKIP absence or downregulation and its clinical
significance in a variety of human cancers, such as prostate,
breast and gastric cancers (12–14). These
observations led to intensive studies on the mechanisms and
functions of RKIP in cancers; however, few previous studies have
analyzed the RKIP methylation and expression status in ESCC. Using
immunohistochemical analysis, Kim et al demonstrated that
71.4% of ESCC metastatic lymph nodes, 50.0% of primary ESCC and
28.9% of carcinoma in situ showed the loss of RKIP
expression, suggesting that RKIP plays an inhibitory role in the
invasion and metastasis of ESCC (15). In addition, Gao et al reported
that positive expression of RKIP in ESCC occurred significantly
less often than in paratumor normal tissues, and that reduced RKIP
expression was significantly correlated with a higher risk of
recurrence, implying its importance in prognosis prediction
(16). Furthermore, the expression
level of RKIP was decreased in the order of dysplastic Barrett's
mucosa, low-grade dysplasia, high-grade dysplasia and EAC,
suggesting its involvement in esophageal carcinogenesis (17).
Although these studies all support the significance
of RKIP in ESCC, minimal information is known regarding the
mechanisms leading to RKIP silencing in ESCC. Considering that
promoter hypermethylation is an important mechanism for gene
silencing, and that it plays an important role in downregulating
RKIP in gastric carcinoma (18,19), the
authors of the present study hypothesized that promoter methylation
may be a potential mechanism for downregulating RKIP in ESCC and is
thus responsible for the biological behaviors of ESCC. To test this
hypothesis, the RKIP promoter methylation status and its expression
level in ESCC tissues and matched paratumor normal tissues were
examined using methylation-specific polymerase chain reaction (MSP)
and immunohistochemical staining, respectively. Furthermore, their
correlations with distinct clinicopathological features of the
patients were analyzed.
Materials and methods
Patients and tissue samples
The present study was approved by the Ethics
Committee of Hebei Medical University (Shijiazhuang, China), where
all data were collected, and written informed consent was obtained
from all participants. A total of 77 patients (59 males and 18
females; mean age of 61.8±8.7 years) who underwent radical surgery
of ESCC in the Fourth Hospital of Hebei Medical University between
December 2008 and December 2010 were recruited to this study. No
patients received pre-operative chemotherapy or radiotherapy. Among
these patients, 29 were aged <60 years, while 49 were aged
>60 years. According to the tumor, lymph node, metastasis (TNM)
staging criteria from the Union for International Cancer Control
and the American Joint Committee on Cancer (20), 33 cases were of stage I or II and 44
cases were of stage III or IV. According to the pathological
grading system (21), 20 ESCC cases
were well-differentiated, 26 were moderately-differentiated, and 31
were poorly-differentiated cancers. Among these ESCC cases, 45
showed positive lymph node metastasis, while 32 showed negative
lymph node metastasis. The clinicopathological characteristics of
the patients are summarized in Table
I.
 | Table I.Clinicopathological characteristics
of esophageal squamous cell carcinoma cases in this study. |
Table I.
Clinicopathological characteristics
of esophageal squamous cell carcinoma cases in this study.
| Characteristic | n | % |
|---|
| Gender |
|
|
|
Male | 59 | 76.62 |
|
Female | 18 | 23.38 |
| TNM stage |
|
|
| I +
II | 33 | 42.86 |
| III+
IV | 44 | 57.14 |
| Pathological
differentiation |
|
|
|
Well | 20 | 25.97 |
|
Moderate | 26 | 33.77 |
|
Poor | 31 | 40.26 |
| Age (years) |
|
|
|
<60 | 29 | 37.66 |
|
≥60 | 48 | 62.34 |
| Lymph node
metastasis |
|
|
|
Negative | 32 | 42.86 |
|
Positive | 45 | 57.14 |
During surgery, the cancer tissues and the matched
normal tissues were obtained from the primary tumors and at least
5-cm away from the tumor, respectively. All tissues were
immediately stored at −80°C, with some subsequently used for DNA
extraction and some fixed in 10% formalin and embedded in paraffin
for immunohistochemical staining. All matched normal tissues were
confirmed by pathological examination to contain no invaded tumor
cells.
Modified MSP method
Tissue samples were treated with proteinase K, and
genomic DNA was extracted using phenol/chloroform. Using the
TU-1800 PC UV-VIS spectrophotometer (Beijing, China) to measure the
absorbance ratio at 260/280 nm, the total DNA purity was determined
to be between 1.8 and 2.0. Bisulfite modification of 10 µg genomic
DNA from each sample was performed as described previously
(22). Briefly, genomic DNA was
denatured using 2 M NaOH and then incubated with 10 M hydroquinone
(Merck Millipore, Darmstadt, Germany) and 3 M sodium bisulfite at
50°C for 16 h. Modified DNA was purified using the Wizard DNA
purification resin (Promega Corporation, Madison, WI, USA),
according to the manufacturer's protocol. The MSP was performed
using primers (Table II) under the
following conditions: Predenaturation at 95°C for 12 min; 36 cycles
at 95°C for 60 sec, 52°C for 45 sec and 72°C for 60 sec; and a
final extension at 72°C for 10 min. The polymerase chain reaction
(PCR) products were subsequently separated on 2% agarose gels by
electrophoresis and analyzed using an ultraviolet light gel imaging
system (FOTODYNE Incorporated, Hartland, WI, USA). The peripheral
blood from a healthy individual without any diseases or tumors in
the alimentary system was taken to isolate genomic DNA, which,
following methylation treatment using DNA methyltransferase (Sss I;
Beijing Solarbio Science & Technology, Co., Ltd., Beijing,
China) was used as a positive control; that without Sss I treatment
was used as a negative control. A water blank was also used as a
negative control. The tissue samples showing positive PCR products
only with methylated primers (full methylation) or with both
methylated and unmethylated primers (semi-methylation) were defined
as methylated samples (23).
 | Table II.Sequences, Tm and amplicon sizes of
primers used for RKIP methylation-specific PCR. |
Table II.
Sequences, Tm and amplicon sizes of
primers used for RKIP methylation-specific PCR.
|
| Primer sequences,
5′-3′ | Tm, °C | PCR product size,
bp |
|---|
| M | F
TTTAGCGATATTTTTTGAGATACGA | 52.5 | 205 |
|
| R
GCTCCCTAACCTCTAATTAACCG |
|
|
| U | F
TTTAGTGATATTTTTTGAGATATGA | 52.5 | 205 |
|
| R
CACTCCCTAACCTCTAATTAACCAA |
|
|
Immunohistochemical staining
Immunohistochemical staining for RKIP was performed
on 4-µm formalin-fixed, paraffin-embedded tissue sections using the
Immunohistochemical Streptavidin-Peroxidase kit (ZSGB-BIO, Beijing,
China), according to the manufacturer's protocol. Briefly, the
tissue sections were dewaxed, dehydrated and treated with 3%
hydrogen peroxidase to block endogenous peroxidase activity.
Following antigen retrieval in EDTA solution (pH 8.5) in a pressure
cooker for 3 min, the sections were blocked with 10% normal goat
serum at 37°C for 40 min, followed by incubation with the primary
rabbit anti-human RKIP antibody (catalog no., bs-1436R; dilution,
1:500; Bioss, Beijing, China) and then horseradish
peroxidase-labeled streptavidin. Final color development was
achieved using a 3,3′-diaminobenzidine solution, and the slides
were counterstained with 1% Meyer's hematoxylin. The use of
phosphate-buffered saline instead of the primary antibody served as
a negative control.
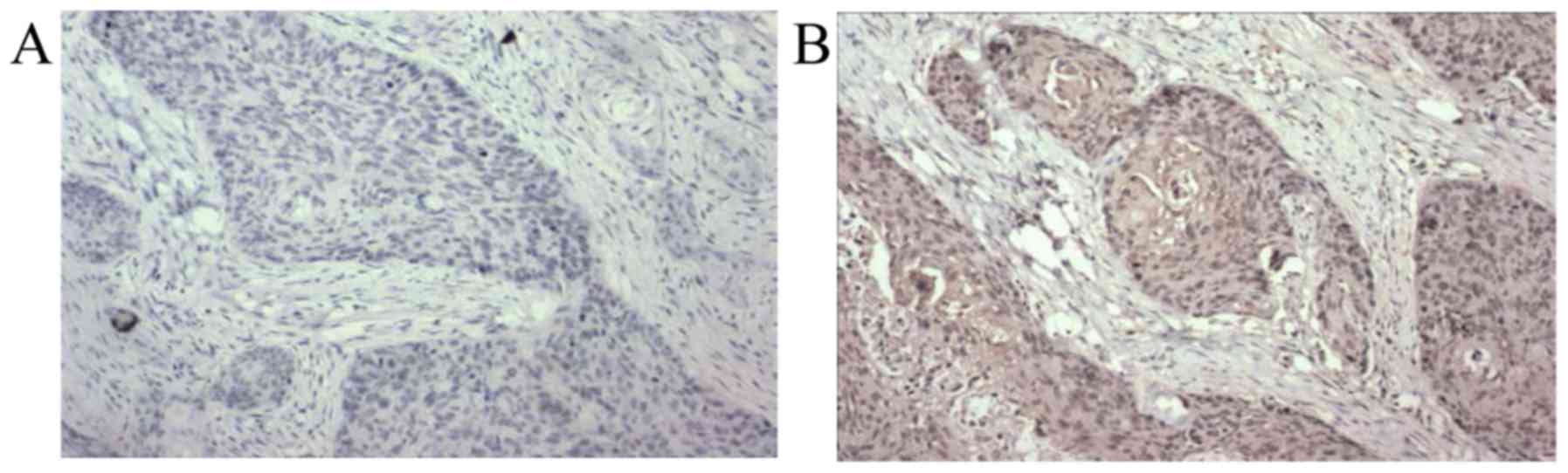
Positive RKIP staining appeared as yellowish to
brownish granules in the cytoplasm. The staining was scored and
averaged by three independent clinical pathologists according to a
scoring system modified from that proposed by Fromowitz et
al (24). Briefly, five random
fields were imaged from each slide under a BX41 light microscope
(Olympus, Tokyo, Japan). A score was assigned based on the
percentage of positive tumor cells averaged from all five fields:
Score 0, ≤25%; score 1, 26–50%; score 2, 51–75%; and score 3,
>75%. In addition, the staining was graded based on the
intensity of the majority of the positively stained cells: 0, no
staining; 1, weak yellowish staining; 2, moderate brownish
staining; and 3, dark brownish staining. The final score was
obtained by adding the percentage score and intensity grade, and
stratified as: ‘-’=0; ‘+’=1–2; ‘++’=3–4; and ‘+++’=5–6, where ‘-’
and ‘+’ were classified as negative expression, while ‘++’ and
‘+++’ were classified as positive expression.
Statistical analysis
All statistical analyses were performed using SPSS
software version 13.0 (SPSS, Inc., Chicago, IL, USA). Quantitative
data are presented as a percentage or ratio of the total sample.
The association between groups was assessed using the χ2
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
RKIP promoter methylation and protein
expression in ESCC and paratumor normal tissues
To examine the status of RKIP promoter methylation
in ESCC, MSP analysis on 77 ESCC tumor tissues and matched
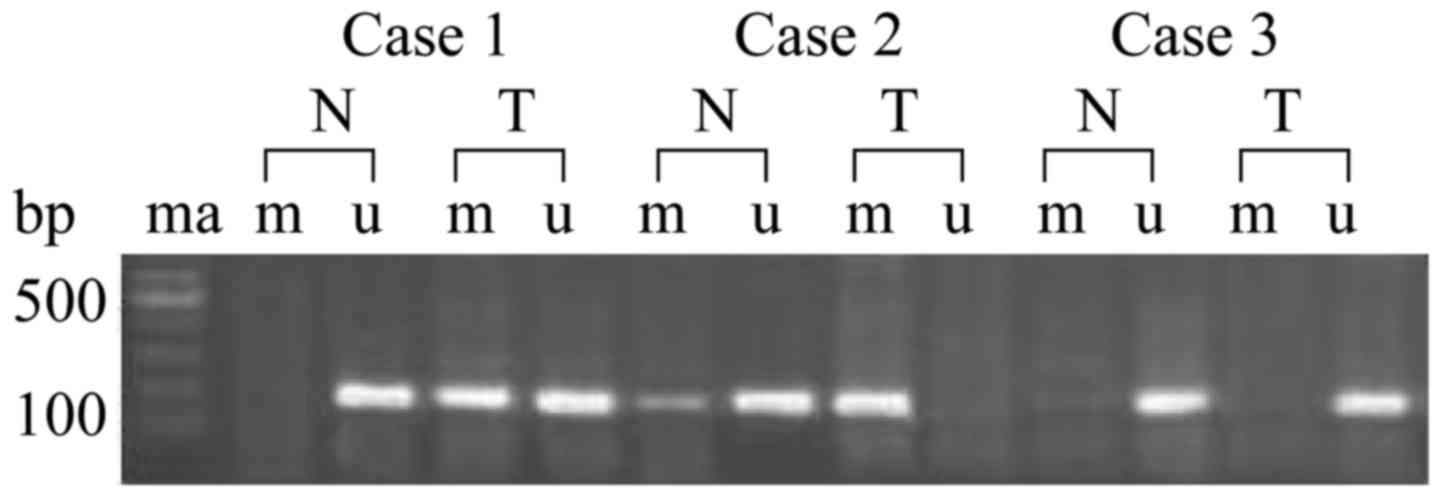
paratumor normal tissues was performed (Fig. 1). The incidence of RKIP promoter
methylation in ESCC tissues was 75.3%, which was significantly
higher compared with the matched normal tissues (27.3%; P<0.001;
Table III). Given that
methylation-induced gene silencing is an important mechanism for
downregulating many tumor suppressor genes during cancer
development (25,26), the expression level of RKIP in ESCC
and normal tissues was also profiled by immunohistochemistry
(Fig. 2). The incidence of positive
RKIP expression in the ESCC tissues was 36.4%, which was
significantly reduced compared with the matched normal tissues
(76.6%; P<0.001; Table III).
 | Table III.Methylation status and protein
expression of RKIP in 77 ESCC tissues and the matched paratumor
normal tissues. |
Table III.
Methylation status and protein
expression of RKIP in 77 ESCC tissues and the matched paratumor
normal tissues.
|
| RKIP promoter
methylation | RKIP protein
expression |
|---|
|
|
|
|
|---|
| Tissues | Methylation | No methylation | Positive | Negative |
|---|
| ESCC (n=77) | 58 (75.32%) | 19 (24.67%) | 28 (36.36%) | 49 (63.64%) |
| Normal (n=77) | 21 (27.27%) | 54 (72.73%) | 59 (76.62%) | 18 (23.38%) |
| χ2 | 35.582 | 25.389 |
| P-value | P<0.001 | P<0.001 |
Correlation between RKIP promoter
methylation and the clinicopathological characteristics of
ESCC
The correlation between RKIP promoter methylation in
ESCC and various clinicopathological characteristics is shown in
Table IV. The incidence of RKIP
promoter methylation was not significantly correlated with the age
(P=0.528), gender (P=1.000) or TNM stage (P=0.321), although it was
significantly correlated with the differentiation status of the
tumor (50.0% in well-differentiated tumors, 73.1% in
moderately-differentiated tumors and 93.5% in poorly differentiated
tumors; P=0.002), as well as the lymph node status (86.7 and 59.4%
in ESCC patients with and without positive lymph node metastasis,
respectively; P=0.006).
 | Table IV.Correlation between RKIP promoter
methylation and the clinicopathological characteristics. |
Table IV.
Correlation between RKIP promoter
methylation and the clinicopathological characteristics.
|
| RKIP promoter
methylation |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | M | U | χ2 | P-value |
|---|
| Age (years) |
|
|
|
| 0.528 |
|
<60 | 29 | 23 | 6 | 0.398 |
|
|
≥60 | 48 | 35 | 13 |
|
|
| Gender |
|
|
|
| 1.000 |
|
Male | 59 | 44 | 15 | 0.076 |
|
|
Female | 18 | 14 | 4 |
|
|
| Clinical stage |
|
|
|
| 0.321 |
|
I+II | 33 | 23 | 10 | 0.984 |
|
|
III+IV | 44 | 35 | 9 |
|
|
| Degree of
differentiation |
|
|
|
| 0.002 |
|
Well | 20 | 10 | 10 | 12.511 |
|
|
Moderate | 26 | 19 | 7 |
|
|
|
Poor | 31 | 29 | 2 |
|
|
| Lymph node
metastasis |
|
|
|
| 0.006 |
|
Negative | 32 | 19 | 13 | 7.494 |
|
|
Positive | 45 | 39 | 6 |
|
|
Correlation between RKIP protein
expression and the clinicopathological characteristics of ESCC
The analysis of the correlation between the
incidence of RKIP protein expression in ESCC and various
clinicopathological characteristics showed that the expression of
RKIP in the tumor tissues was only significantly correlated with
lymph node metastasis (24.4 and 53.1% in ESCC patients with and
without positive lymph node metastasis, respectively; P=0.01); it
was not significantly associated with age (P=0.540), gender
(P=0.760), TNM stage (P=0.056) or the differentiation status of the
tumor (P=0.282; Table V).
 | Table V.Correlation between RKIP protein
expression and clinicopathological characteristics. |
Table V.
Correlation between RKIP protein
expression and clinicopathological characteristics.
|
|
| RKIP protein
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | n | Positive | Negative | χ2 | P-value |
|---|
| Age, years |
|
|
| 0.571 | 0.450 |
|
<60 | 29 | 9 | 20 |
|
|
|
≥60 | 48 | 19 | 29 |
|
|
| Gender |
|
|
| 0.093 | 0.760 |
|
Male | 59 | 22 | 37 |
|
|
|
Female | 18 | 6 | 12 |
|
|
| Clinical stage |
|
|
| 3.667 | 0.056 |
| I +
II | 33 | 16 | 17 |
|
|
| III +
IV | 44 | 12 | 32 |
|
|
| Degree of
differentiation |
|
|
| 2.535 | 0.282 |
|
Well | 20 | 9 | 11 |
|
|
|
Moderate | 26 | 11 | 15 |
|
|
|
Poor | 31 | 8 | 23 |
|
|
| Lymph node
metastasis |
|
|
| 6.648 | 0.010 |
|
Negative | 32 | 17 | 15 |
|
|
|
Positive | 45 | 11 | 34 |
|
|
Association between RKIP promoter
methylation and protein expression
Among the 58 ESCC tissues positive for RKIP promoter
methylation, 29.3% were positive for RKIP protein expression. Of
the 19 ESCC tissues negative for RKIP promoter methylation, 57.9%
were positive for RKIP protein expression. There was a significant
negative association between RKIP promoter methylation and protein
expression (P=0.001; Table VI).
 | Table VI.Association between RKIP promoter
methylation and protein expression in esophageal squamous cell
carcinoma tissues. |
Table VI.
Association between RKIP promoter
methylation and protein expression in esophageal squamous cell
carcinoma tissues.
|
| RKIP protein
expression |
|
|
|
|---|
|
|
|
|
|
|
|---|
| RKIP methylation
status | + | – | Total | χ2 | P-value |
|---|
| Methylated | 17 | 41 | 58 | 17.308 | 0.001 |
| Unmethylated | 11 | 8 | 19 |
|
|
| Total | 28 | 49 | 77 |
|
|
Discussion
The present study examined the status of RKIP
promoter methylation and its expression in ESCC and matched
paratumor normal tissues, and demonstrated that RKIP promoter
methylation was significantly enhanced, while its protein
expression level was significantly reduced, in the tumor tissues
compared with the normal tissues. Functionally, RKIP promoter
methylation was significantly correlated with the status of tumor
differentiation and lymph node metastasis, while RKIP protein
expression was associated with lymph node metastasis only.
Consistent with these observations, RKIP promoter methylation was
negatively associated with its protein expression in ESCC
tissues.
Tumorigenesis is a complicated process involving
numerous factors, multiple steps and genetic as well as epigenetic
alterations in various genes. The pathogenesis of ESCC has been
mainly attributed to environmental factors, including malnutrition,
smoking, alcohol use and inflammation (27–29).
However, recent studies have revealed that genetic and epigenetic
alterations also play significant roles in the development of
multiple cancers, including ESCC (30–32). Among
various epigenetic alterations, aberrant DNA methylation (including
hypomethylation of oncogenes and hypermethylation of tumor
suppressor genes) is the best characterized and most crucial
mechanism modulating chromatin structure and the expression levels
of oncogenes or tumor suppressor genes, contributing to tumor
initiation and further development (33). Hypermethylation of the CpG islands
within the promoter regions of tumor suppressor genes is a
well-demonstrated molecular mechanism for transcriptional silencing
and subsequent tumor progression (34). The presence of promoter CpG island
hypermethylation in preneoplastic lesions of malignant cancers
supports its significance in neoplastic transformation and tumor
initiation (35–38).
Many studies have corroborated the nature of RKIP as
a tumor suppressor gene. Li et al reported that promoter
hypermethylation of the RKIP gene led to its silencing in
colorectal cancer, which may underlie tumor metastasis (39). Al-Mulla et al demonstrated that
chromosome loss in colorectal cancer was inversely proportional to
RKIP expression levels, the silencing of which was mainly caused by
methylation of the RKIP promoter (40). Consistently, other studies have
supported the importance of RKIP promoter hypermethylation in the
loss of its activity (41,42). In contrast, few studies have
investigated the status or clinical significance of RKIP promoter
methylation in ESCC. To address this issue, the present study
compared promoter methylation of the RKIP gene in ESCC tissues and
the matched normal tissues of 77 patients with ESCC, and showed
that RKIP promoter hypermethylation was significantly enhanced in
the tumor tissues compared with the normal tissues, supporting its
potential involvement in tumor initiation. Furthermore, RKIP
promoter hypermethylation was significantly correlated with tumor
differentiation and lymph node metastasis, suggesting its value in
predicting ESCC metastasis and prognosis.
In the paratumor normal tissues, 27.3% were positive
for RKIP promoter methylation, and all these cases were
semi-methylated (data not shown). One potential explanation for
this was that we used the highly sensitive nested MSP for this
study, which is capable of detecting alleles comprising <5% of
the total genomic DNA (43).
Therefore, minor invasion of the tumor tissue into the normal
tissue (although negative in the pathological examinations) may
have led to semi-methylation. Among the 58 ESCC samples positive
for RKIP promoter methylation, 3 were semi-methylated (data not
shown), which may be related to the degree of transcriptional
silencing of RKIP; that is, an incomplete methylation corresponds
to incomplete gene silencing (44).
In addition to promoter methylation, reduced or loss
of RKIP expression has been associated with cancer development.
Immunohistochemical analysis has demonstrated that the loss of RKIP
expression is a key phenotype for many human cancers, including
colorectal cancer, breast cancer, melanoma, prostate cancer and
hepatocellular carcinoma, modulating distant metastasis, lymphatic
metastasis, vascular infiltration and cancer mortality (45–49).
Consistent with these studies, the present study showed that the
incidence of positive RKIP expression in ESCC tissues was
significantly less compared with the matched normal tissues, and
that it was clinically correlated with lymph node metastasis. A
further association analysis revealed that promoter
methylation-induced RKIP silencing may be a major mechanism for
downregulating RKIP and subsequent ESCC development.
The potential involvement of RKIP as a tumor
suppressor gene in tumor invasiveness and metastasis has been well
demonstrated in multiple cancers. Keller et al reported that
RKIP expression was at its highest in normal prostate tissues,
decreased in primary prostate cancers and was not detectable in
metastatic tissues from prostate cancer (50). In addition, Schuierer et al
demonstrated that RKIP expression was decreased concomitantly with
the metastasis of melanoma (47).
Mechanistic studies have suggested that RKIP is a metastasis
suppressor gene that is responsible for blocking several signaling
pathways in the metastatic cascade, including MEK, G proteins and
NF-κB (51). Similarly, the present
study showed that the percentage of RKIP expression in ESCC tumors
with lymph node metastasis was significantly less compared with
tumors without lymph node metastasis.
In summary, this study showed that promoter
methylation may be responsible for RKIP downregulation and the
oncogenesis of ESCC. Therefore, the incidence of RKIP promoter
methylation may serve as a biomarker for the differentiation status
and prognosis of ESCC. This study extends the molecular
understanding of the pathogenesis of ESCC and provides a novel
target for the early diagnosis and treatment of ESCC.
Acknowledgements
This study was supported by the Research Project of
Science and Technology of Handan (grant no. 1123108079-4).
References
|
1
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li H: Modern esophageal surgery. People's
Military Medical Press; Beijing: pp. 331–332. 2004
|
|
3
|
Esophageal cancer, . Epidemiology,
pathogenesis and prevention. Nat Clin Pract Gastroenterol Hepatol.
5:517–526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holmes RS and Vaughan TL: Epidemiology and
pathogenesis of esophageal cancer. Semin Radiat Oncol. 17:2–9.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li JS, Ying JM, Wang XW, Wang ZH, Tao Q
and Li LL: Promoter methylation of tumor suppressor genes in
esophageal squamous cell carcinoma. Chin J Cancer. 32:3–11. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng CP, Kuo IY, Alakus H, Frazer KA,
Harismendy O, Wang YC and Tseng VS: Network-based analysis
identifies epigenetic biomarkers of esophageal squamous cell
carcinoma progression. Bioinformatics. 30:3054–3061. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bernier I and Jollés P: Purification and
characterization of a basic 23 kDa cytosolic protein from bovine
brain. Biochim Biophys Acta. 790:174–181. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yeung K, Seitz T, Li S, Janosch P,
McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, et
al: Suppression of Raf-1 kinase activity and MAP kinase signalling
by RKIP. Nature. 401:173–177. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yeung KC, Rose DW, Dhillon AS, Yaros D,
Gustafsson M, Chatterjee D, McFerran B, Wyche J, Kolch W and Sedivy
JM: Raf kinase inhibitor protein interacts with NF-kappaB-inducing
kinase and TAK1 and inhibits NF-kappaB activation. Mol Cell Biol.
21:7207–7217. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deiss K, Kisker C, Lohse MJ and Lorenz K:
Raf kinase inhibitor protein (RKIP) dimer formation controls its
target switch from Raf1 to G protein-coupled receptor kinase (GRK)
2. J Biol Chem. 287:23407–23417. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Escara-Wilke J, Yeung K and Keller ET: Raf
kinase inhibitor protein (RKIP) in cancer. Cancer Metastasis Rev.
31:615–620. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu Z, Smith PC, Zhang L, Rubin MA, Dunn
RL, Yao Z and Keller ET: Effects of raf kinase inhibitor protein
expression on suppression of prostate cancer metastasis. J Natl
Cancer Inst. 95:878–889. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Z, Lu P, Zhang H, Xu H, Gao N, Li M
and Liu C: Nestin positively regulates the Wnt/β-catenin pathway
and the proliferation, survival and invasiveness of breast cancer
stem cells. Breast Cancer Res. 16:4082014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu C, Lu P, Lu Y, Xu H, Wang S and Chen
J: Clinical implications of metastatic lymph node ratio in gastric
cancer. BMC Cancer. 7:2002007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim HS, Won KY, Kim GY, Kim SC, Park YK
and Kim YW: Reduced expression of Raf-1 kinase inhibitory protein
predicts regional lymph node metastasis and shorter survival in
esophageal squamous cell carcinoma. Pathol Res Pract. 208:292–299.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao C, Pang L, Ren C and Ma T: Prognostic
value of raf kinase inhibitor protein in esophageal squamous cell
carcinoma. Pathol Oncol Res. 18:471–477. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Birner P, Jesch B, Schultheis A and
Schoppmann SF: RAF-kinase inhibitor protein (RKIP) downregulation
in esophageal cancer and its metastases. Clin Exp Metastasis.
29:551–559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo W, Dong Z, Guo Y, Lin X, Chen Z, Kuang
G and Yang Z: Aberrant methylation and loss expression of RKIP is
associated with tumor progression and poor prognosis in gastric
cardia adenocarcinoma. Clin Exp Metastasis. 30:265–275. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li DX, Cai HY, Wang X, Feng YL and Cai SW:
Promoter methylation of Raf kinase inhibitory protein: A
significant prognostic indicator for patients with gastric
adenocarcinoma. Exp Ther Med. 8:844–850. 2014.PubMed/NCBI
|
|
20
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rice TW, Blackstone EH and Rusch VW: 7th
edition of the AJCC Cancer Staging Manual: Esophagus and
esophagogastric junction. Ann Surg Oncol. 17:1721–1724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Glickman JF, Flynn J and Reich NO:
Purification and characterization of recombinant
baculovirus-expressed mouse DNA methyltransferase. Biochem Biophys
Res Commun. 230:280–284. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fromowitz FB, Viola MV, Chao S, Oravez S,
Mishriki Y, Finkel G, Grimson R and Lundy J: ras p21 expression in
the progression of breast cancer. Hum Pathol. 18:1268–1275. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2:(Suppl 1). S4–S11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010.PubMed/NCBI
|
|
27
|
Yang CS: Vitamin nutrition and
gastroesophageal cancer. J Nutr. 130:(Suppl 2S). S338–S339.
2000.
|
|
28
|
Yokokawa Y, Ohta S, Hou J, Zhang XL, Li
SS, Ping YM and Nakajima T: Ecological study on the risks of
esophageal cancer in Ci-Xian, China: The importance of nutritional
status and the use of well water. Int J Cancer. 83:620–624. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Launoy G, Milan CH, Faivre J, Pienkowski
P, Milan CI and Gignoux M: Alcohol, tobacco and oesophageal cancer:
Effects of the duration of consumption, mean intake and current and
former consumption. Br J Cancer. 75:1389–1396. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun
ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al: Genetic landscape of
esophageal squamous cell carcinoma. Nat Genet. 46:1097–1102. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sasaki Y, Tamura M, Koyama R, Nakagaki T,
Adachi Y and Tokino T: Genomic characterization of esophageal
squamous cell carcinoma: Insights from next-generation sequencing.
World J Gastroenterol. 22:2284–2293. 2016.PubMed/NCBI
|
|
32
|
Kailasam A, Mittal SK and Agrawal DK:
Epigenetics in the pathogenesis of esophageal adenocarcinoma. Clin
Transl Sci. 8:394–402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma K, Cao B and Guo M: The detective,
prognostic, and predictive value of DNA methylation in human
esophageal squamous cell carcinoma. Clin Epigenetics. 8:432016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shames DS, Minna JD and Gazdar AF: DNA
methylation in health, disease, and cancer. Curr Mol Med. 7:85–102.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jones PA, Rideout WM III, Shen JC, Spruck
CH and Tsai YC: Methylation, mutation and cancer. Bioessays.
14:33–36. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pogribny I, Raiche J, Slovack M and
Kovalchuk O: Dose-dependence, sex- and tissue-specificity, and
persistence of radiation-induced genomic DNA methylation changes.
Biochem Biophys Res Commun. 320:1253–1261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brooks JD, Weinstein M, Lin X, Sun Y, Pin
SS, Bova GS, Epstein JI, Isaacs WB and Nelson WG: CG island
methylation changes near the GSTP1 gene in prostatic
intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev.
7:531–536. 1998.PubMed/NCBI
|
|
38
|
Chan AO and Rashid A: CpG island
methylation in precursors of gastrointestinal malignancies. Curr
Mol Med. 6:401–408. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li FF, Song SJ and Zhang RN:
Phosphatidylethanolamine-binding protein (PEBP) in basic and
clinical study. Sheng Li Ke Xue Jin Zhan. 40:214–218. 2009.(In
Chinese). PubMed/NCBI
|
|
40
|
Al-Mulla F, Hagan S, Al-Ali W, Jacob SP,
Behbehani AI, Bitar MS, Dallol A and Kolch W: Raf kinase inhibitor
protein: Mechanism of loss of expression and association with
genomic instability. J Clin Pathol. 61:524–529. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Eves EM, Shapiro P, Naik K, Klein UR,
Trakul N and Rosner MR: Raf kinase inhibitory protein regulates
aurora B kinase and the spindle checkpoint. Mol Cell. 23:561–574.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Martinho O, Gouveia A, Silva P, Pimenta A,
Reis RM and Lopes JM: Loss of RKIP expression is associated with
poor survival in GISTs. Virchows Arch. 455:277–284. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Licchesi JD and Herman JG:
Methylation-specific PCR. Methods Mol Biol. 507:305–323. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bird A: Molecular biology. Methylation
talk between histones and DNA. Science. 294:2113–2115. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Minoo P, Zlobec I, Baker K, Tornillo L,
Terracciano L, Jass JR and Lugli A: Loss of raf-1 kinase inhibitor
protein expression is associated with tumor progression and
metastasis in colorectal cancer. Am J Clin Pathol. 127:820–827.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hagan S, Al-Mulla F, Mallon E, Oien K,
Ferrier R, Gusterson B, García JJ and Kolch W: Reduction of Raf-1
kinase inhibitor protein expression correlates with breast cancer
metastasis. Clin Cancer Res. 11:7392–7397. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schuierer MM, Bataille F, Hagan S, Kolch W
and Bosserhoff AK: Reduction in Raf kinase inhibitor protein
expression is associated with increased Ras-extracellular
signal-regulated kinase signaling in melanoma cell lines. Cancer
Res. 64:5186–5192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fu Z, Kitagawa Y, Shen R, Shah R, Mehra R,
Rhodes D, Keller PJ, Mizokami A, Dunn R, Chinnaiyan AM, et al:
Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a
novel prognostic marker in prostate cancer. Prostate. 66:248–256.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schuierer MM, Bataille F, Weiss TS,
Hellerbrand C and Bosserhoff AK: Raf kinase inhibitor protein is
downregulated in hepatocellular carcinoma. Oncol Rep. 16:451–456.
2006.PubMed/NCBI
|
|
50
|
Keller ET, Fu Z, Yeung K and Brennan M:
Raf kinase inhibitor protein: A prostate cancer metastasis
suppressor gene. Cancer Lett. 207:131–137. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Keller ET: Metastasis suppressor genes: A
role for raf kinase inhibitor protein (RKIP). Anticancer Drugs.
15:663–669. 2004. View Article : Google Scholar : PubMed/NCBI
|