Introduction
Esophageal squamous cell carcinoma (ESCC) is the
major histological type of esophageal cancer in East Asian
countries, and is the eighth most common malignancy, and the sixth
leading cause of cancer death worldwide (1). Despite improvements in surgical
procedures, multidisciplinary therapies and perioperative
management, the prognosis of patients with ESCC remains
unsatisfactory (2). A staging system
for ESCC based on clinicopathological features that can provide an
accurate prediction of survival in patients aids clinicians in
selecting the best treatment approach. Currently, the TNM
classification is widely used to stage patients and to select
treatment strategies (3). However,
the assessment of prognosis using this system remains inadequate
due to the considerable biological heterogeneity within the same
stage of ESCC. Therefore, novel biological markers are required
that provide a more accurate prognosis of patients and that
facilitate in selecting the appropriate treatment strategy for
individual patients.
Heat-shock transcription factor 1 (HSF1) is a
transcription factor that induces the expression of heat shock
proteins (Hsp) in response to various stressors (4,5). These
Hsps act as molecular chaperones to maintain cellular homeostasis
by restoring protein folding and stability. HSF1 facilitates
survival in response stressors by regulating various cellular
processes, such as cell-cycle control, protein translation and
glucose metabolism (6,7). The expression of HSF1 is reportedly
upregulated in cancer cell lines and cancer tissues of
hepatocellular carcinoma, and high expression of HSF1 is associated
with a poor prognosis in breast cancer and hepatocellular carcinoma
(8,9).
However, the role of HSF1 as a prognostic biomarker in ESCC has not
been well investigated. Therefore, the present study analyzed HSF1
mRNA and protein expression in resected ESCC tissues to determine
if the expression of HSF1 could be used as a prognostic biomarker
in ESCC.
Materials and methods
Sample collection
Samples of resected ESCC and normal squamous
epithelium were obtained from two independent cohorts, and assayed
for HSF1 mRNA and protein expression. Cohort 1 was composed of 58
ESCC samples, which were collected during subtotal esophagectomy
between February 2005 and March 2009 after obtaining written
informed consent. All patients underwent subtotal esophagectomy at
one of five hospitals (Juntendo University Hospital, Tokyo, Japan;
National Cancer Center Hospital, Tokyo, Japan; Kurume University
Hospital, Fukuoka, Japan; Saitama Cancer Center, Saitama, Japan;
Kagoshima University Hospital, Kagoshima, Japan). The
clinicopathological characteristics of patients are indicated in
Table I. All patients underwent
curative resection and no patients received neoadjuvant therapy.
The tumor samples were submitted for cDNA microarray analysis.
 | Table I.Correlation between HSF1 mRNA
expression and clinicopathological parameters in cohort 1. |
Table I.
Correlation between HSF1 mRNA
expression and clinicopathological parameters in cohort 1.
| Parameter | Low (n=29) | High (n=29) | P-value |
|---|
| Age,
yearsa | 66 (52–82) | 66 (52–79) | 0.72 |
| Gender, n |
|
|
|
| Male | 24 | 18 | 0.08 |
|
Female | 5 | 1 |
|
| Location, n |
|
|
|
| Ut | 3 | 8 | 0.24 |
| Mt | 16 | 13 |
|
| Lt | 10 | 8 |
|
| Histological grade,
n |
|
|
|
| G1 | 12 | 8 | 0.49 |
| G2 | 14 | 16 |
|
| G3 | 3 | 5 |
|
| pT stage, n |
|
|
|
| 1 | 4 | 4 | 0.69 |
| 2 | 3 | 5 |
|
| 3 | 19 | 15 |
|
| 4 | 3 | 5 |
|
| pN stage, n |
|
|
|
| 0 | 15 | 9 | 0.61 |
| 1 | 5 | 6 |
|
| 2 | 6 | 5 |
|
| 3 | 3 | 9 |
|
Cohort 2 consisted of 212 ECC samples collected
during subtotal esophagectomy at Osaka University Hospital (Osaka,
Japan) between June 2000 and February 2013, following receipt of
written informed consent. Patient characteristics, including the
number of patients that underwent neoadjuvant therapy, are
indicated in Table II. These
specimens were submitted for immunohistochemical analysis. The
present study was approved by the Ethics Committee of Osaka
University Hospital (Osaka, Japan) and the written consent of all
the patients was obtained. Pathological tumor stage was evaluated
using the seventh edition of the TNM classification established by
the Union for International Cancer Control (3).
 | Table II.Correlation between HSF1 protein
expression and clinicopathological parameters in cohort 2. |
Table II.
Correlation between HSF1 protein
expression and clinicopathological parameters in cohort 2.
| Parameter | Low expression
(n=103) | High expression
(n=109) | P-value |
|---|
| Age,
yearsa | 64 (47–79) | 65 (39–80) | 0.43 |
| Gender, n |
|
|
|
|
Male | 89 | 95 | 0.87 |
|
Female | 14 | 14 |
|
| Histological grade,
n |
|
|
|
| G1 | 27 | 22 | 0.14 |
| G2 | 43 | 62 |
|
| G3 | 24 | 16 |
|
| Gx | 9 | 9 |
|
| Location, n |
|
|
|
| Ut | 11 | 18 | 0.03 |
| Mt | 65 | 47 |
|
| Lt | 27 | 44 |
|
| pT stage, n |
|
|
|
| T1 | 33 | 24 | 0.41 |
| T2 | 16 | 19 |
|
| T3 | 51 | 61 |
|
| T4 | 3 | 5 |
|
| pN stage, n |
|
|
|
| N0 | 41 | 31 | 0.12 |
| N1 | 37 | 35 |
|
| N2 | 16 | 29 |
|
| N3 | 9 | 13 |
|
| pStage, n |
|
|
|
| I | 22 | 18 | 0.10 |
| II | 34 | 24 |
|
|
III | 31 | 49 |
|
| IV | 16 | 18 |
|
| Neoadjuvant
therapy |
|
|
|
|
None | 39 | 46 | 0.52 |
|
Chemotherapy | 64 | 63 |
|
Extraction of RNA and cDNA microarray
analysis
Resected tumor tissues in cohort 1 were immediately
cut (5 mm thickness), and embedded in Tissue-Tek OCT medium (Sakura
Finetek Europe B.V., Alphen aan den Rijn, The Netherlands), frozen
in liquid nitrogen and maintained at −80°C until RNA extraction.
Following isolation of RNA and DNA using the QIAamp DNA Micro kit
(Qiagen, Valencia, CA, USA) and the RNeasy Micro kit (Qiagen), cDNA
was synthesized from 8.0 µg of total RNA (10) using the commercially available Whole
Human Genome Oligo DNA Microarray kit (Agilent Technologies, Inc.,
Santa Clara, CA, USA). Cyanine-labeled cRNA was prepared using T7
linear amplification as described in the Agilent Low RNA Input
Fluorescent Linear Amplification kit manual (Agilent Technologies,
Inc.). Labeled cRNA was then fragmented (with fragmentation buffer
from the Agilent Low RNA Input Fluorescent Amplification kit,
according to the manufacturer's protocol; Agilent Technologies,
Inc.) and hybridized to an oligonucleotide microarray (Whole Human
Genome Microarray kit, 4×44K; product no. G4112F; Agilent
Technologies, Inc.). Fluorescence intensities were determined with
a DNA microarray scanner (G2565BA/DB; Agilent Technologies, Inc.)
and analyzed using G2567AA Feature Extraction Software (version
A7.5.1; Agilent Technologies, Inc.), which used the locally
weighted linear regression curve fit normalization method (11). This microarray study followed Minimum
Information About a Microarray Experiment guidelines issued by the
Microarray Gene Expression Data group (12). Further analyses were performed using
GeneSpring (version 7.3; Agilent Technologies, Inc.). mRNA
expression level in cancer tissue by microarray was categorised by
the median expression level into high and low expression
groups.
Immunohistochemistry
Tumor specimens in cohort 2 were fixed in 10%
formalin (Wako Pure Chemical Industries, Ltd., Osaka, Japan) and
embedded in paraffin (Wako Pure Chemical Industries, Ltd.). The
expression of proteins was evaluated by immunohistochemistry in
4-µm-thick sections following deparaffinization with xylene (Wako
Pure Chemical Industries, Ltd.) and dehydration in graded ethanol
solutions. For antigen retrieval, the sections were autoclaved in
10 mM citrate buffer (pH 6.0) at 115°C for 20 min. After blocking
with hydrogen peroxide (Wako Pure Chemical Industries, Ltd.), the
sections were incubated with specific antibodies overnight at 4°C
in a moist chamber. Rat monoclonal antibody against HSF1 (10H8;
Enzo Life Sciences, Inc., Farmingdale, NY, USA), mouse monoclonal
antibody against Hsp27 (G3.1; Enzo Life Sciences, Inc.), mouse
monoclonal antibody against Hsp70 (C92F3A-5; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and mouse monoclonal antibody
against Hsp90 (ADI-SPA-830-D; Enzo Life Sciences, Inc.), were used
at dilutions of 1:100, 1:1,000, 1:500 and 1:300, respectively.
Sites of antibody binding were visualized with the ABC peroxidase
detection system according to the manufacturer's protocol (catalog
nos., PK-4004 and PK-4010; Vector Laboratories, Inc., Burlingame,
CA, USA) using a fluorescence microscope (BZ-X700; Keyence
Corporation, Osaka, Japan). The intensity of antibody-stained
nuclei in cancer cells was evaluated by two independent
pathologists, who were blinded to the clinicopathological
parameters. The intensity of HSF1, Hsp27, Hsp70 and Hsp90
immunoreactions in the cells were scored as follows: 0, not
stained; 1+, weak staining; 2+, moderate staining; and 3+, intense
staining. Finally, the expression of these proteins in ESCC
specimens was classified into a HSF1 low expression group (0 or 1+)
and a HSF1 high expression group (2+ or 3+).
Statistical analysis
The association between HSF1 protein expression and
various clinicopathological factors was analyzed using the χ2 test
and Fisher's exact probability test. The overall survival (OS) was
assessed by performing the Kaplan-Meier method and compared using
the log-rank test. OS was defined as the period from the date of
surgery to the date of death from any cause. All parameters with a
P<0.10 in univariate analysis by the Cox proportional hazard
model were entered into multivariate survival analysis. P<0.05
was considered to indicate a statistically significant difference.
Statistical analysis was performed with JMP Pro software (version
10.0.2; SAS Institute, Cary, NC, USA). Each test was repeated six
times.
Results
Correlation between HSF1 mRNA
expression and clinicopathological characteristics in cohort 1
The 58 ESCC cases were classified into two groups
using the median HSF1 mRNA expression level in cancer tissue as
determined by microarray. Table I
shows the correlation between HSF1 expression and various
clinicopathological parameters. No significant association was
observed with age, gender, tumor location, histological grade,
pathological T (pT) stage or pathological N (pN) stage
(P<0.05).
Correlation between HSF1 mRNA
expression and OS in cohort 1
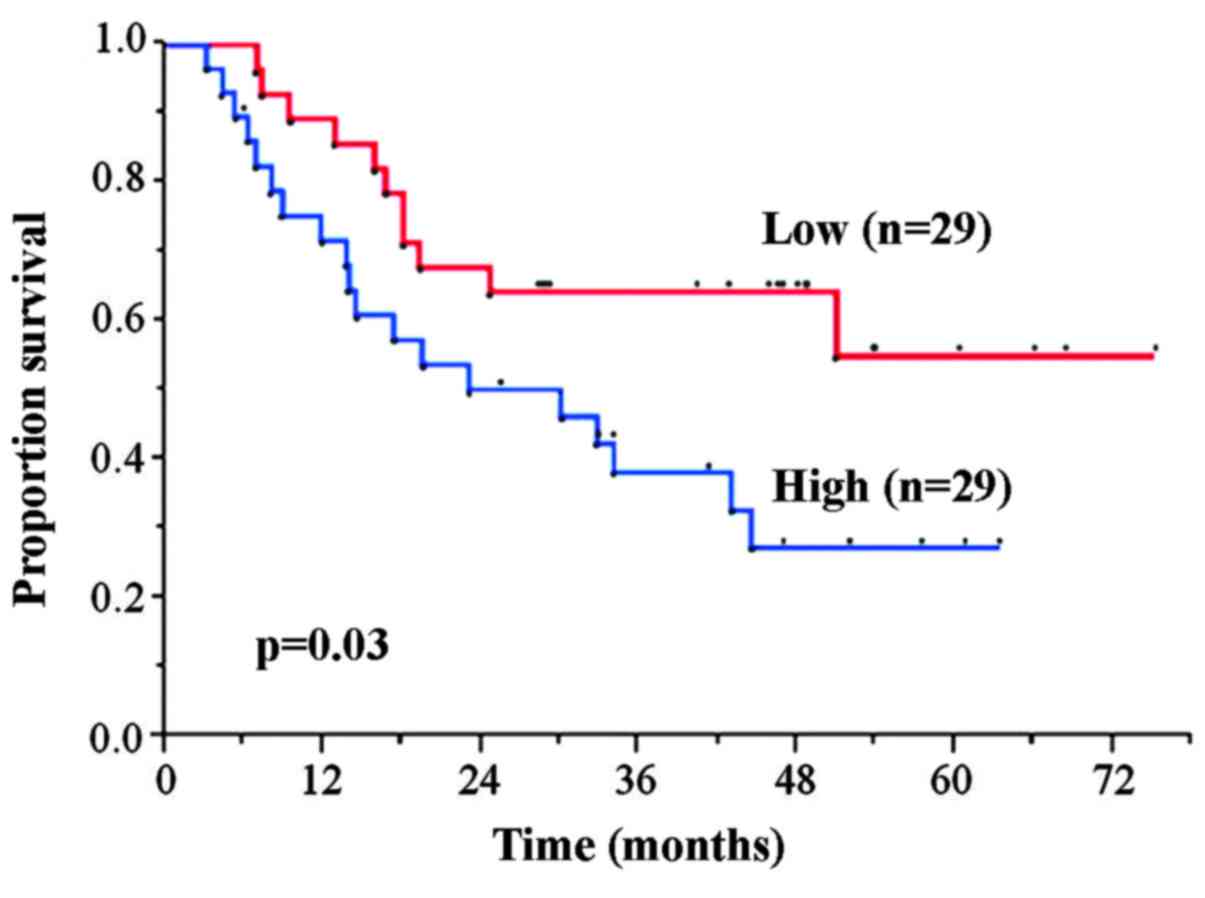
The 5-year OS rates in the HSF1 high and low
expression groups was 27.3 and 55.1%, respectively. The HSF1 high
expression group exhibited significantly worse OS rate compared
with the low expression group (P=0.03) (Fig. 1).
HSF1 protein expression in cohort
2
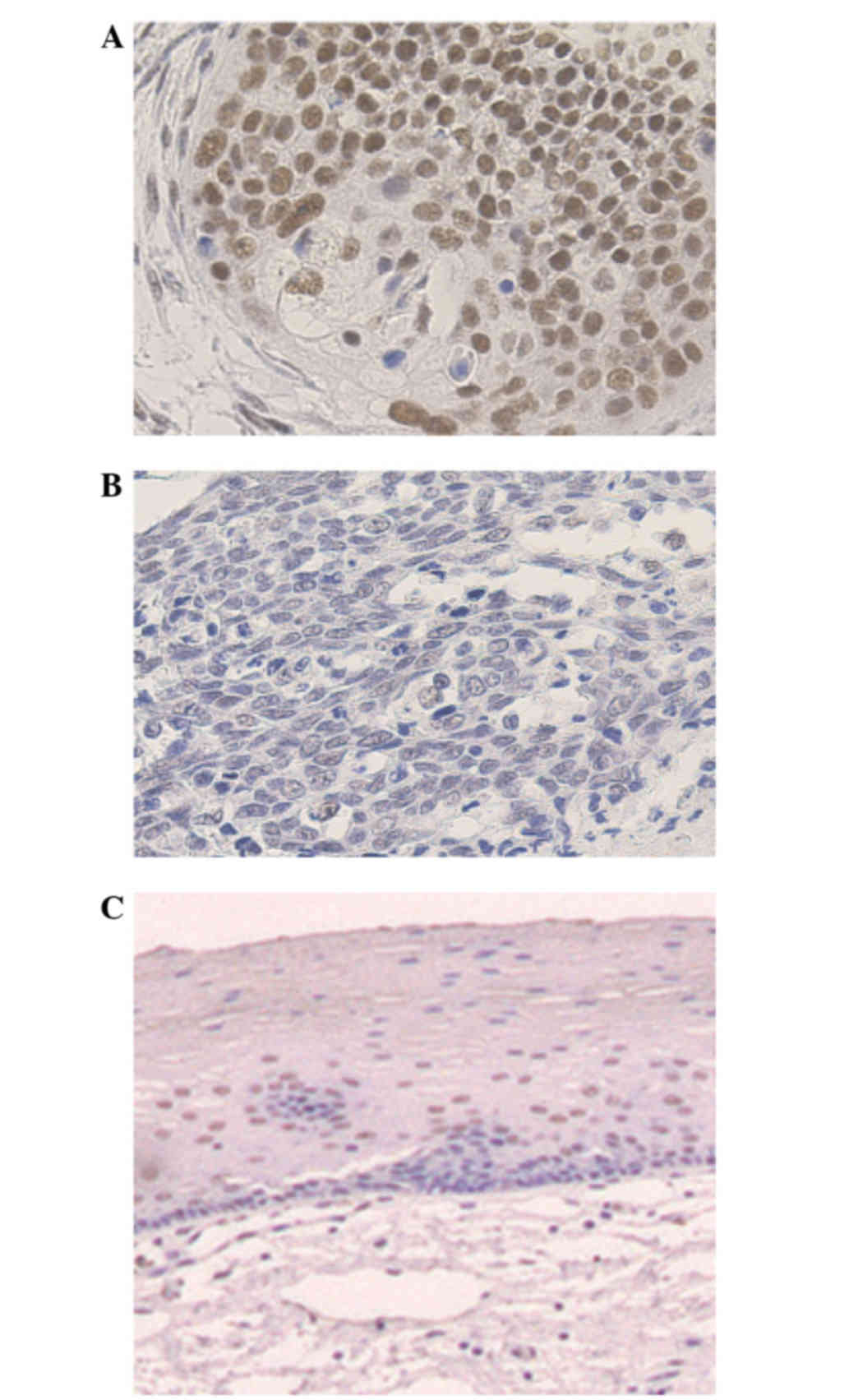
A total of 212 cancerous lesion samples were
evaluated for HSF1 protein expression by immunohistochemical
analysis. Of these, 109 (51.4%) exhibited high HSF1 expression
(Table II), predominantly in the
nuclei of the tumor cells, with faint cytoplasmic staining
(Fig. 2A), while the remaining 103
(48.6%) samples showed low HSF1 expression (Fig. 2B). The staining was almost homogenous
among different areas of the cancerous lesions, including the
surface, central and deep areas. None of the normal squamous
epithelium samples showed significant levels of HSF1 expression,
although faint immunostaining was observed in the nuclei of a small
number of basal cells (Fig. 2C).
Scoring of the immunostained sections was almost identical by the
two pathologists, with interobserver variation of <5%.
Correlation between HSF1 protein
expression and clinicopathological parameters in cohort 2
Table II indicates
the correlations between HSF1 protein expression and various
clinicopathological parameters. In comparison to primary tumors
with low HSF1 expression, primary tumors with high HSF1 expression
were significantly more likely to be in the upper location
(P=0.03). All other parameters, including age, gender, pT, pN,
pathological stage and preoperative therapy, did not significantly
correlate with HSF1 protein expression.
Correlation between HSF1 protein
expression and clinical outcome in cohort 2
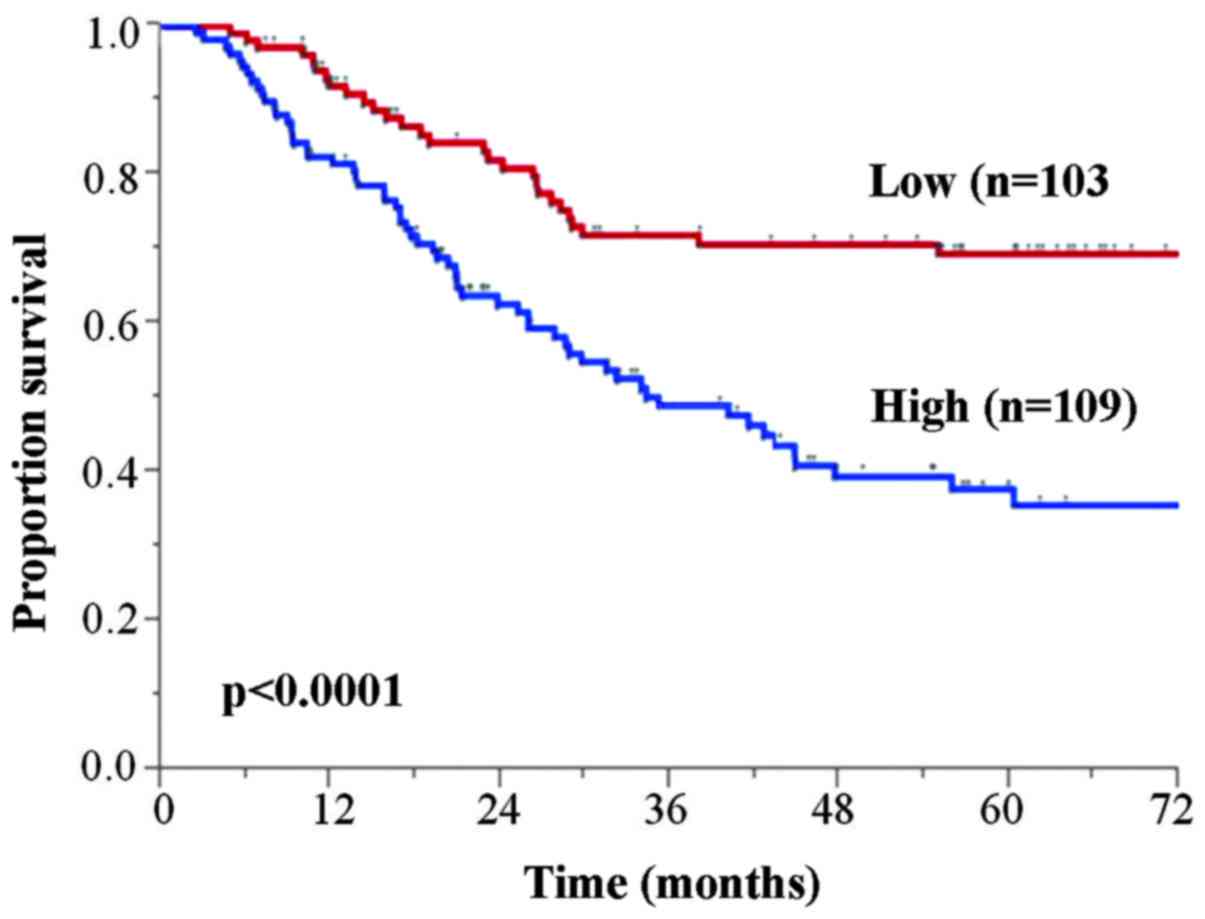
The 5-year OS rates for patients with low and high
HSF1 expression tumors were 69.4 and 37.8%, respectively (Fig. 3). The high HSF1 expression group
exhibited significantly poorer OS than those in the low HSF1
expression group (P<0.0001). Furthermore, univariate analysis
revealed that the OS significantly correlated with age, pT, pN and
HSF1 protein expression (Table
III). Multivariate analysis using the five parameters with
P<0.10 on univariate analysis identified high HSF1 expression
[hazard ratio (HR), 2.29; 95% confidence interval (CI), 1.48–3.64;
P=0.0002] as an independent OS prognostic factor, in addition to pT
(HR, 2.21; 95% CI, 1.38–3.65; P=0.0008) and pN (HR, 1.73; 95% CI,
1.04–3.02; P=0.03).
 | Table III.Univariate and multivariate survival
analyses of overall survival rate by Cox's proportional hazard
model. |
Table III.
Univariate and multivariate survival
analyses of overall survival rate by Cox's proportional hazard
model.
|
|
| Univariate | Multivariate |
|---|
|
|
|
|
|
|---|
| Parameter | Patients, n | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years
(65≤/≤64) | 106/106 | 1.64 | 1.08–2.50 | 0.02 | 1.51 | 0.99–2.32 | 0.06 |
| Gender
(male/female) | 184/28 | 1.36 | 0.74–2.8 | 0.34 |
|
|
|
| Histological grade
(G1/G2, G3, Gx) | 49/163 | 0.81 | 0.51–1.32 | 0.39 |
|
|
|
| Location (Ut,
Mt/Lt) | 141/71 | 0.66 | 0.44–1.02 | 0.06 | 1.36 | 0.88–2.07 | 0.16 |
| pT (T3, T4/T1,
T2) | 92/120 | 2.66 | 1.69–4.34 | <0.0001 | 2.22 | 1.39–3.65 | 0.0007 |
| pN (N1, N2,
N3/N0) | 72/140 | 2.42 | 1.49–4.14 | 0.0002 | 1.76 | 1.06–3.06 | 0.03 |
| Neoadjuvant therapy
(none/chemotherapy) | 85/127 | 0.91 | 0.59–1.38 | 0.65 |
|
|
|
| HSF1 expression
(high/low) | 109/103 | 2.53 | 1.64–4.0 | <0.0001 | 2.24 | 1.44–3.56 | 0.0003 |
Correlation between HSF1 protein
expression and expression of Hsps in cohort 2
To determine whether the expression of Hsps
correlated with HSF1 tumor expression in ESCC tumor specimens,
55/212 ESCC randomly selected samples in cohort 2 were
immunohistochemically evaluated for Hsp27, Hsp70 and Hsp90
expression. High expression of HSF1 and Hsp27 was observed in 24
ESCC samples, 18 samples showed low expression of HSF1 and Hsp27, 7
samples exhibited high expression of HSF1 but low expression of
Hsp27, and 6 samples exhibited low HSF1 expression but high Hsp27
expression (Table IV). The results
indicate that HSF1 expression is significantly correlated with
Hsp27 expression (P<0.0001). Hsp90 expression was also
significantly correlated with HSF1 expression (P<0.0001), but
Hsp70 expression was not (P=0.38).
 | Table IV.Correlation between HSF1, and Hsp27,
Hsp70 and Hsp90 expression. |
Table IV.
Correlation between HSF1, and Hsp27,
Hsp70 and Hsp90 expression.
|
| HSF1 |
|
|
|---|
|
|
|
|
|
|---|
| Expression | + | − | Total | P-value |
|---|
| Hsp27 |
|
|
|
|
| + | 24 | 6 | 30 | <0.0001 |
| − | 7 | 18 | 25 |
|
| Hsp70 |
|
|
|
|
| + | 7 | 8 | 15 | 0.38 |
| − | 24 | 16 | 40 |
|
| Hsp90 |
|
|
|
|
| + | 28 | 7 | 35 | <0.0001 |
| − | 3 | 17 | 20 |
|
| Total | 31 | 24 | 55 |
|
These results indicate that the expression of HSF1
is closely correlated with poor prognosis in ESCC. The results also
indicate that Hsp27 and Hsp90 expression, but not Hsp70 expression,
are possible downstream targets of HSF1 in ESCC.
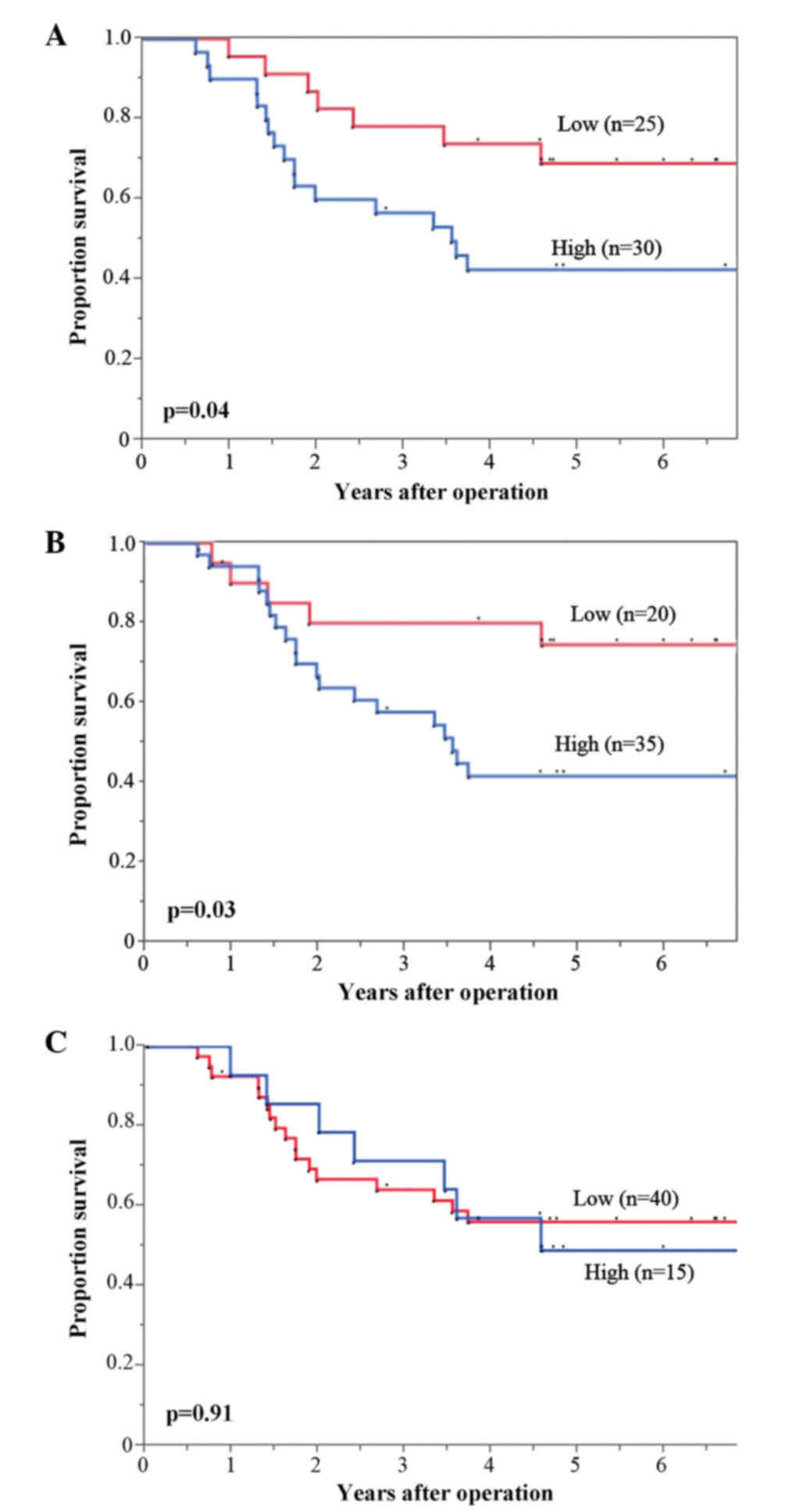
Thus, the correlation between the Hsp27, Hsp70 and
Hsp90 expression, and OS were investigated. The high Hsp27 and
Hsp90 expression groups showed significantly poorer OS compared
with those in the low Hsp27 and Hsp90 expression groups (P=0.04 and
P=0.03, respectively) (Fig. 4A and B,
respectively), while there were no significant differences in OS
between high and low Hsp70 expression (P=0.91) (Fig. 4C).
Discussion
Hsps are overexpressed in numerous types of cancer
(including hepatocellular carcinoma, colorectal carcinoma, bladder
cancer and neuroblastoma), and are correlated with tumor
proliferation, invasion and metastasis (13,14).
Various studies report that Hsp levels are useful biomarkers of
carcinogenesis and aggressiveness in specific types of cancer,
including hepatocellular carcinoma, colorectal carcinoma, breast
cancer, primary oral squamous cell carcinoma and gastric
adenocarcinoma (15–18). Hsp expression is regulated by HSF1,
which can bind to the 5′ promoter regions of all Hsp genes, and
trigger their immediate and massive transcription (19,20). HSF1
expression itself is also involved in numerous crucial steps of
carcinogenesis and tumor development (7). Its expression is closely correlated with
invasion, metastasis and angiogenesis (21,22), and
has been identified as a prognostic factor in breast cancer and
hepatocellular carcinoma.
The present study demonstrated that high HSF1
expression is associated with poor prognosis in two independent
patient cohorts, and appears to be an independent prognostic factor
for patients undergoing curative resection for ESCC. The prediction
of prognosis following curative surgical resection will enable
clinicians to determine the necessity for intensive follow-up and
adjuvant therapy (23,24). Furthermore, if HSF1 expression can be
measured in pretreated biopsy samples it may yield useful
information for determining the treatment strategy for individual
patients.
Additionally, the present study revealed that the
expression of Hsp27 and Hsp90 are closely correlated with HSF1
expression in ESCC tumor specimens. This suggests that the
HSF1/Hsp27 and HSF1/Hsp90 pathways may be involved in the
progression of ESCC. A number of studies previously reported that
Hsp27 and Hsp90 were associated with survival and could be
potential molecular targets in various types of cancer (23,25–29).
Recently, Hsp90 inhibitors have been developed, and
used as preclinical and clinical anticancer therapies (30,31). The
present study indicates that Hsp27 and Hsp90 may be potential
therapeutic targets in patients with ESCC, as HSF1 is a regulator
of Hsps. Therefore, inhibiting HSF leads to inhibition of not only
Hsp27, but also Hsp90. However, the results suggest that HSF1,
which regulates the transcription of Hsp27 and Hsp90, may be a more
effective and stronger therapeutic target. HSF1 may, therefore, be
a valid prognostic marker in ESCC and a candidate for novel
approaches to ESCC treatment.
Prospective studies are required to further clarify
the significance of HSF1 in the clinical setting. Additionally, it
will be important to investigate the molecular mechanisms
underlying the effect of HSF1 expression on prognosis. The present
findings may mark a new step for the exploration of appropriate
treatment strategies and the development of novel therapeutic
approaches for ESCC.
In conclusion, the results of the present study
indicate that HSF1 is a prognostic factor for patients with ESCC,
and that Hsp27 and Hsp90, but not Hsp70, may be the downstream
targets of HSF1 in ESCC.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fujita H: The history of lymphadenectomy
for esophageal cancer and the future prospects for esophageal
cancer surgery. Surg Today. 45:140–149. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sobin LH, Gospodarowicz MK and Wittekind
Ch: Oesophagus including oesophagogastric junctionTNM
Classification of Malignant Tumors. 7th. Wiley-Blackwell; Oxford:
pp. 65–72. 2009
|
|
4
|
Sorger PK: Heat shock factor and the heat
shock response. Cell. 65:363–366. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qian SB, McDonough H, Boellmann F, Cyr DM
and Patterson C: CHIP-mediated stress recovery by sequential
ubiquitination of substrates and Hsp70. Nature. 440:551–555. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Page TJ, Sikder D, Yang L, Pluta L,
Wolfinger RD, Kodadek T and Thomas RS: Genome-wide analysis of
human HSF1 signaling reveals a transcriptional program linked to
cellular adaptation and survival. Mol Biosyst. 2:627–639. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dai C, Whitesell L, Rogers AB and
Lindquist S: Heat shock factor 1 is a powerful multifaceted
modifier of carcinogenesis. Cell. 130:1005–1018. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li S, Ma W, Fei T, Lou Q, Zhang Y, Cui X,
Qin X, Zhang J, Liu G, Dong Z, et al: Upregulation of heat shock
factor 1 transcription activity is associated with hepatocellular
carcinoma progression. Mol Med Rep. 10:2313–2321. 2014.PubMed/NCBI
|
|
9
|
Santagata S, Hu R, Lin NU, Mendillo ML,
Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM,
Lindquist S and Ince TA: High levels of nuclear heat-shock factor 1
(HSF1) are associated with poor prognosis in breast cancer. Proc
Natl Acad Sci USA. 108:18378–18383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Enders GH: Cyclins in breast cancer: Too
much of a good thing. Breast Cancer Res. 4:145–147. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Quackenbush J: Microarray data
normalization and transformation. Nat Genet. 32:(Suppl). S496–S501.
2002. View
Article : Google Scholar
|
|
12
|
Brazma A, Hingamp P, Quackenbush J,
Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA,
Causton HC, et al: Minimum information about a microarray
experiment (MIAME)-toward standards for microarray data. Nat Genet.
29:365–371. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khalil AA, Kabapy NF, Deraz SF and Smith
C: Heat shock proteins in oncology: Diagnostic biomarkers or
therapeutic targets? Biochim Biophys Acta. 1816:89–104.
2011.PubMed/NCBI
|
|
14
|
Ciocca DR and Calderwood SK: Heat shock
proteins in cancer: Diagnostic, prognostic, predictive and
treatment implications. Cell Stress Chaperones. 10:86–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kaigorodova EV and Bogatyuk MV: Heat shock
proteins as prognostic markers of cancer. Curr Cancer Drug Targets.
14:713–726. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ciocca DR and Vargas-Roig LM: Hsp27 as a
prognostic and predictive factor in cancer. Prog Mol Subcell Biol.
28:205–218. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim KK, Jang TJ and Kim JR: HSP70 and ER
expression in cervical intraepithelial neoplasia and cervical
cancer. J Korean Med Sci. 13:383–388. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chuma M, Sakamoto M, Yamazaki K, Ohta T,
Ohki M, Asaka M and Hirohashi S: Expression profiling in multistage
hepatocarcinogenesis: Identification of HSP70 as a molecular marker
of early hepatocellular carcinoma. Hepatology. 37:198–207. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu C: Heat shock transcription factors:
Structure and regulation. Annu Rev Cell Dev Biol. 11:441–469. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Calderwood SK, Xie Y, Wang X, Khaleque MA,
Chou SD, Murshid A, Prince T and Zhang Y: Signal transduction
pathways leading to heat shock transcription. Sign Transduct
Insights. 2:13–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khaleque MA, Bharti A, Gong J, Gray PJ,
Sachdev V, Ciocca DR, Stati A, Fanelli M and Calderwood SK: Heat
shock factor 1 represses estrogen-dependent transcription through
association with MTA1. Oncogene. 27:1886–1893. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gabai VL, Meng L, Kim G, Mills TA,
Benjamin IJ and Sherman MY: Heat shock transcription factor Hsf1 is
involved in tumor progression via regulation of hypoxia-inducible
factor 1 and RNA-binding protein HuR. Mol Cell Biol. 32:929–940.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lyu X, Huang J, Mao Y, Liu Y, Feng Q, Shao
K, Gao S, Jiang Y, Wang J and He J: Adjuvant chemotherapy after
esophagectomy: Is there a role in the treatment of the lymph node
positive thoracic esophageal squamous cell carcinoma? J Surg Oncol.
110:864–868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ruiz MI Gallegos, Floor K, Roepman P,
Rodriguez JA, Meijer GA, Mooi WJ, Jassem E, Niklinski J, Muley T,
van Zandwijk N, et al: Integration of gene dosage and gene
expression in non-small cell lung cancer, identification of HSP90
as potential target. PLoS One. 3:e00017222008.PubMed/NCBI
|
|
25
|
Nakamura M, Iwahashi M, Nakamori M, Ojima
T, Katsuda M, Iida T, Hayata K, Kato T and Yamaue H: New prognostic
score for the survival of patients with esophageal squamous cell
carcinoma. Surg Today. 44:875–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pick E, Kluger Y, Giltnane JM, Moeder C,
Camp RL, Rimm DL and Kluger HM: High HSP90 expression is associated
with decreased survival in breast cancer. Cancer Res. 67:2932–2937.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Solit DB, Scher HI and Rosen N: Hsp90 as a
therapeutic target in prostate cancer. Semin Oncol. 30:709–716.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Romani AA, Crafa P, Desenzani S, Graiani
G, Lagrasta C, Sianesi M, Soliani P and Borghetti AF: The
expression of HSP27 is associated with poor clinical outcome in
intrahepatic cholangiocarcinoma. BMC Cancer. 7:2322007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bauer K, Nitsche U, Slotta-Huspenina J,
Drecoll E, von Weyhern CH, Rosenberg R, Höfler H and Langer R: High
HSP27 and HSP70 expression levels are independent adverse
prognostic factors in primary resected colon cancer. Cell Oncol
(Dordr). 35:197–205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smyth T, Van Looy T, Curry JE,
Rodriguez-Lopez AM, Wozniak A, Zhu M, Donsky R, Morgan JG, Mayeda
M, Fletcher JA, et al: The HSP90 inhibitor, AT13387, is effective
against imatinib-sensitive and -resistant gastrointestinal stromal
tumor models. Mot Cancer Ther. 11:1799–1808. 2012. View Article : Google Scholar
|
|
31
|
Wagner AJ, Agulnik M, Heinrich MC,
Mahadevan D, Riedel RF, von Mehren M, Trent J, Demetri GD, Corless
CL and Yule M: Dose-escalation study of a second-generation
non-ansamycin HSP90 inhibitor, onalespib (AT13387), in combination
with imatinib in patients with metastatic gastrointestinal stromal
tumour. Eur J Cancer. 61:94–101. 2016. View Article : Google Scholar : PubMed/NCBI
|