Introduction
Breast cancer is the most frequent cancer affecting
females, and the leading cause of cancer-related mortality among
females worldwide (1). Although the
disease has been studied widely, its underlying molecular
mechanisms have not been fully elucidated. microRNAs (miRNAs) are a
class of non-coding small RNAs (~22 nucleotides) which negatively
regulate the expression of target messenger RNAs (mRNAs) by binding
to their 3′-untranslated regions (3′UTRs), causing mRNA degradation
and/or translation inhibition (2).
miRNAs have been implicated in a variety of biological processes,
including embryonic development, cell differentiation and diseases
including human cancer (3). miR-198
has been reported to be deregulated in several human cancers,
including colorectal (4), lung
(5), pancreatic (6,7),
hepatocellular (8,9), prostate (10) and esophageal cancer (11). However, the involvement and effects of
miR-198 on breast cancer progression and the underlying mechanism
remain unknown.
CUB domain-containing protein 1 (CDCP1) has been
widely reported to be highly expressed in various human cancers,
and is significantly correlated with tumor malignancy and poor
prognosis (12–17). CDCP1 is a transmembrane protein with
several conserved tyrosine residues in the cytoplasmic domain that
may be phosphorylated by the Src family kinases (18,19).
Previous studies have indicated that CDCP1 is involved in
tumorigenesis processes by regulating cell migration ability and
matrix degradation in a tyrosine phosphorylation-dependent manner
(17). In breast cancer, tumors with
a high level of CDCP1 expression demonstrate higher levels of
proliferation (20), and CDCP1 is
also suggested to be responsible for the regulation of adhesion and
motility in breast cancer cells (21). These results indicate that CDCP1 may
play a critical role in human breast cancer progression. However,
the molecular mechanisms underlying CDCP1 regulation in human
breast cancer cells have not been fully elucidated. Our
computational prediction revealed that the CDCP1 3′-untranslated
region (3′UTR) has miR-198 binding sites, suggesting that CDCP1 may
be a direct target of miR-198.
In the present study, we aimed to elucidate the
involvement of the miR-198/CDCP1 interaction in human breast
cancer. Our results indicated that miR-198 was frequently
downregulated in human breast cancer tissues and cell lines. In
addition, enhanced expression of miR-198 reduced cell proliferation
and migration, and promoted cell adhesion in breast cancer cells
in vitro. Moreover, transcription activator-like effector
nuclease (TALEN)-based CDCP1 silencing inhibited cell proliferation
and migration, and promoted cell adhesion, which was similar to the
effects of overexpression of miR-198. Luciferase reporter assay
further demonstrated that miR-198 directly targeted the 3′UTR of
CDCP1. Thus, we provided evidence to characterize the role of
miR-198 and CDCP1 in human breast cancer, which may be useful for
effective clinical therapies in the future.
Materials and methods
Tissue samples and cell lines
Forty-nine clinical breast tumor tissues and the
adjacent tissues (at least 5 cm away from the primary tumor) were
collected at Xiangya Hospital, Changsha, China. All patients signed
an informed consent form and the study was approved by the
Independent Ethical Committee of Central South University,
Changsha, China. Samples were stored at −80°C until use. Four
aggressive breast cancer cell lines (MCF-7, MDA-MB-231, BT474 and
BT549) and a normal breast cell line (MCF10A) were used. Cells were
grown routinely in HEPES-buffered Dulbecco's modified Eagle's
medium containing 10% fetal bovine serum (Gibco Life Technologies,
Carlsbad, CA, USA) and cultured under 5% CO2 humidified
air.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNAs were prepared using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions. The extracted RNA was
reverse-transcribed to cDNA using a PrimeScript reagent kit
(Promega Corporation, Madison, WI, USA). The relative expression of
CDCP1 mRNA was detected by SYBR-Green qPCR assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) performed on an ABI Prism
7700 (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). β-actin was used as a control to normalize the starting
quantity of RNA. The specific primers were as follows: CDCP1, F:
5′-TCTGCAAGGCTGTGACCAAG-3′, R: 5′-GCTCATTACTCAAGTCAACCAC-3′;
β-actin, F: 5′-AGGGGCCGGACTCGTCATACT-3′, R:
5′-GGCGGCACCACCATGTACCCT-3′. The specific primers sets for miR-198,
U6 and the PCR mix were purchased from GeneCopoeia, Inc.
(Rockville, MD, USA). The expression of U6 was used as an
endogenous control. Reactions for each sample were performed in
triplicate. Relative expression levels were calculated using the
2−∆∆Ct method.
Western blot analysis
Total cellular extracts were prepared from each
group of cells with 200 ml lysis buffer and subjected to western
blot analysis. Approximately 50 µg total protein was separated by
sodium dodecyl sulphate-polyacrylamide gel electrophoresis,
transferred to a polyvinylidene fluoride membrane and incubated
with the indicated antibodies, followed by horseradish
peroxidase-conjugated secondary antibody. Signals were visualized
using enhanced chemiluminescence (ECL) substrates (Millipore,
Billerica, MA, USA). The protein bands were visualized using an ECL
detection kit (GE Healthcare Life Sciences, Chalfont, UK) as
recommended by the manufacturer. β-actin was used for
normalization. Antibodies of CDCP1 and β-actin were obtained from
Abzoom (Abzoom Biolabs, Dallas, TX, USA).
Dual luciferase reporter assay
A fragment of the 3′UTR of CDCP1 containing the
predicted miR-198 target site was amplified and inserted into the
psiCHECK-2 vector (Promega Corporation) downstream of the
luciferase gene sequence. A psiCHECK-2 construct containing the
3′UTR of CDCP1 with a mutant sequence of miR-198 was synthesized.
The wild-type 3′UTR of CDCP1 (Wt-3′UTR of CDCP1) and mutant 3′UTR
of CDCP1 (Mut-3′UTR of CDCP1) primers were as follows: Wt-3′UTR of
CDCP1, F: 5′-CTCGAGGCAAGCCCTGGATTCAGAGT-3′, R:
5′-GCGGCCGCGGATAACCACGAACCGACCTA-3′; Mut-3′UTR of CDCP1, F:
5′-GCGGCCGCGCAAGCCCTGGATTCAGAGT-3′, R:
5′-CTCGAGGGATAACCACGAACCGACCTA-3′. MCF-7 and MDA-MB-231 cells were
plated in 96-well plates, then the Wt-3′UTR of CDCP1-psi-CHECK2 or
the Mut-3′UTR of CDCP1-psi-CHECK2 was co-transfected with
pre-miR-198 and pre-scramble mimics, respectively. The untreated
group was used as a control. Luciferase activity was detected using
a dual-luciferase reporter gene assay kit (Promega Corporation) and
normalized to Renilla activity.
TALEN-mediated knockout of CDCP1
Loss of function is a powerful approach in the study
of gene function. In this study, we used TALEN technology to knock
out the CDCP1 gene in human breast cancer MCF-7 and MDA-MB-231
cells. TALENs designed to target CDCP1 gene were purchased from
Sidansai Biotechnology (Shanghai, China). Cells in 24-well plates
were transfected with 400 ng TALEN expression plasmids using
Lipofectamine 2000 (Invitrogen Life Technologies) according to the
manufacturer's instructions. Western blot analysis was used to
examine the CDCP1 protein expression to validate the efficiency of
TALEN plasmids.
Lentiviral miR-198 infection
Lentiviruses containing miR-198 (Lv-miR-198) or
scramble (Lv-scramble) were purchased from GeneChem (Shanghai,
China). The MCF-7 and MDA-MB-231 cells were cultured to 60–70% of
the plates, and then a concentration of 3×104 TU/well
Lv-miR-198 or Lv-scramble lentivirus was added. RT-qPCR and western
blot analysis were performed to determinate the mRNA and protein
levels of CDCP1 in the MCF-7 and MDA-MB-231 cells after being
infected for 7 days. The cells stably infected with lentivirus were
expanded and harvested for further analysis.
Cell proliferation assay
3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl
tetrazolium bromide (MTT) assay was performed to evaluate cell
proliferation. Briefly, cells were allowed to grow in 96-well
plates with 5,000 cells per well, and incubated for 24, 48 and 72
h, then 10 mg/ml MTT was added to the cells and incubated for 1 h.
The reaction was then terminated by removal of the supernatant, and
200 µl dimethyl sulfoxide was added. After 1 h of incubation, the
optical density at 570 nm of each well was measured with a
microplate reader (Bio-Rad Laboratories, Inc.).
Cell migration assay
Cell migration was determined by Transwell assay.
Cells suspended in serum-free medium were added into the upper
chamber of the insert with Matrigel. Following 24 h incubation at
37°C, cells remaining on the upper side of the membrane were
carefully removed, while cells that had migrated through the
membrane were fixed with 75% alcohol and stained with crystal
violet for 25 min, then washed with water and dried in air. The
imaging and counting of cell numbers were performed using an
inverted microscope (Nikon Corporation, Tokyo, Japan).
Cell adhesion assay
For adhesion assay, cells were seeded in a
Matrigel-coated 96-well-plate. Following incubation for 1 h, the
wells were washed twice with phosphate-buffered saline, fixed in 4%
paraformaldehyde and stained with crystal violet. The imaging and
counting of adherent cells were performed using an inverted
microscope (Nikon Corporation).
Statistical analysis
All data are presented as the mean values ± standard
deviation. Student's t-test was used to analyze the
differences in the experiments. The Chi-squared test was used to
demonstrate the differences in miR-198 or CDCP1 expression with
clinicopathological features. P<0.05 was considered to indicate
a statistically significant difference.
Results
miR-198 is downregulated, while CDCP1
is upregulated, in human breast cancer tissues and cell lines
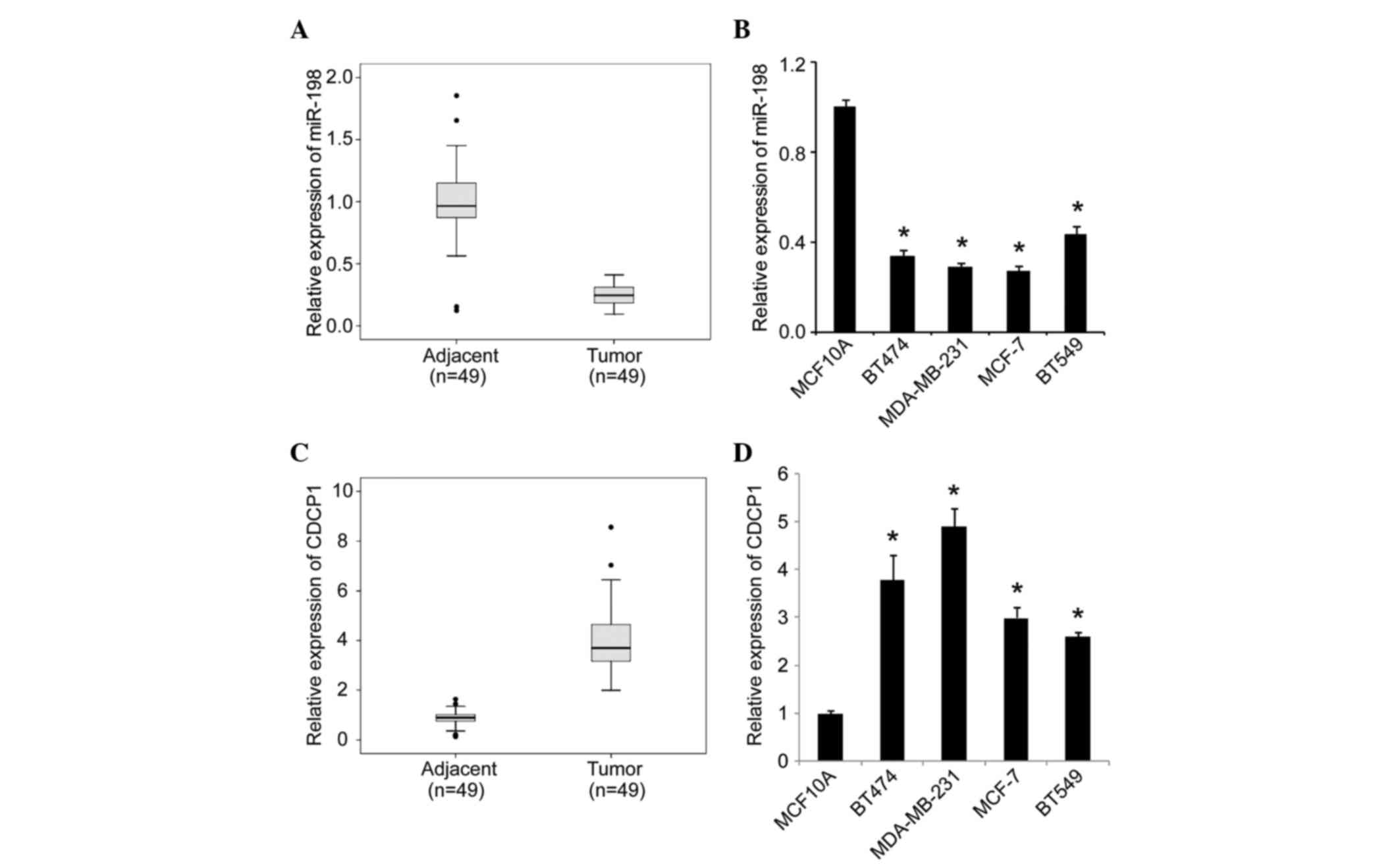
To examine the expression signature of miR-198 in
human breast cancer progression, we first performed miRNA-based
RT-qPCR analysis in 49 clinical tumor samples and matched adjacent
tissues. As shown in Fig. 1A, we
observed that the expression of miR-198 was significantly decreased
in selected tumor tissues compared with the matched adjacent
tissues. Correlation analysis of miR-198 expression with
clinicopathological features revealed that downregulated miR-198
expression was significantly correlated with lymph node metastasis
(P=0.036, Table I). We then moved to
breast cell lines, and observed that the expression of miR-198 was
significantly lower in the four invasive breast cancer cell lines
(BT474, MDA-MB-231, MCF-7 and BT549 cells) than in the normal
breast cell line MCF10A (Fig. 1B).
Thus, our data suggest a strong link between downregulation of
miR-198 and the pathogenesis of breast cancer. Contrary to miR-198,
we observed that CDCP1 was significantly upregulated in selected
clinical tumor samples (Fig. 1C) and
invasive breast cancer cell lines (Fig.
1D). We also analyzed the association of CDCP1 expression with
clinicopathological parameters, and observed that high CDCP1
expression levels were correlated with lymph node metastasis
(P=0.028, Table II). These data
suggest that upregulation of CDCP1 may be involved in breast cancer
progression.
 | Table I.Correlation of miR-198 expression with
clinicopathological features of breast cancer tissues. |
Table I.
Correlation of miR-198 expression with
clinicopathological features of breast cancer tissues.
|
|
| miR-198
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Number of cases | High, n (%) | Low, n (%) | P-value |
|---|
| Age (years) |
|
|
| 0.878 |
| ≤40 | 7 | 2 (28.6) | 5 (71.4) |
|
|
40–50 | 23 | 9 (39.1) | 14 (60.9) |
|
|
50–60 | 13 | 6 (46.2) | 7 (53.8) |
|
| ≥60 | 6 | 2 (33.3) | 4 (66.7) |
|
| TNM classification
(T) |
|
|
| 0.971 |
| T1 | 7 | 3 (42.9) | 4 (57.1) |
|
| T2 | 34 | 13 (38.3) | 21 (61.7) |
|
| T3 | 8 | 3 (37.5) | 5 (62.5) |
|
| TNM classification
(N) |
|
|
| 0.036 |
| N0 | 39 | 18 (46.2) | 21 (53.8) |
|
| N1 | 10 | 1 (10.0) | 9 (90.0) |
|
 | Table II.Correlation of CDCP1 expression with
clinicopathological features of breast cancer tissues. |
Table II.
Correlation of CDCP1 expression with
clinicopathological features of breast cancer tissues.
|
|
| CDCP1 expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Number of
cases | High, n (%) | Low, n (%) | P-value |
|---|
| Age (years) |
|
|
| 0.978 |
|
≤40 | 7 | 4 (57) | 3 (43) |
|
|
40–50 | 23 | 15 (65) | 8 (35) |
|
|
50–60 | 13 | 8 (62) | 5 (38) |
|
|
≥60 | 6 | 4 (67) | 2 (33) |
|
| TNM classification
(T) |
|
|
| 0.404 |
| T1 | 7 | 5 (71) | 2 (29) |
|
| T2 | 34 | 22 (65) | 12 (35) |
|
| T3 | 8 | 7 (88) | 1 (12) |
|
| TNM classification
(N) |
|
|
| 0.028 |
| N0 | 32 | 19 (59) | 13 (41) |
|
| N1 | 10 | 8 (80) | 2 (20) |
|
| N2 | 7 | 7 (100) | 0 (0) |
|
miR-198 directly targets CDCP1 and
inhibits its expression
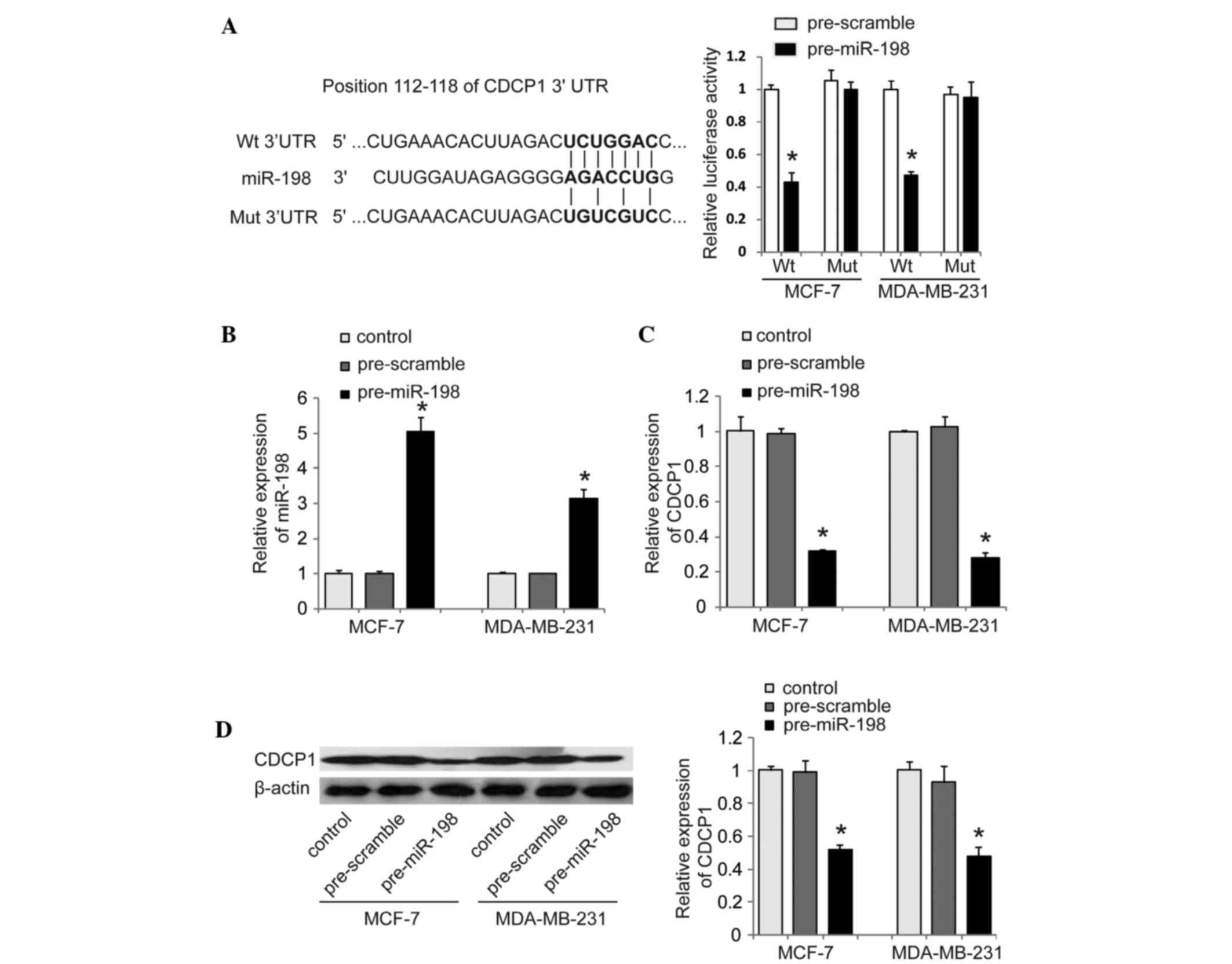
The findings above, as well as the computational
prediction (Fig. 2A) prompted us to
further investigate whether miR-198 directly targets CDCP1. To do
so, we cloned the wild-type 3′UTR (Wt-3′UTR) of CDCP1 containing
the predicted binding site of miR-198 downstream of a luciferase
reporter gene (Fig. 2A). We also
constructed its mutant version (Mut-3′UTR of CDCP1) by binding site
mutagenesis. The vectors were co-transfected with miR-198 mimics
(pre-miR-198) or corresponding scrambled mimics (pre-scramble) as
controls into MCF-7 and MDA-MB-231 cells, respectively. The
luciferase activity of cells transfected with miR-198 mimic was
significantly decreased compared with that of control cells
(Fig. 2A). Additionally, the
miR-198-mediated repression of luciferase activity was abolished by
the mutant putative binding site (Fig.
2A). Furthermore, we tested the inhibitory effect of miR-198 on
CDCP1 expression in MCF-7 and MDA-MB-231 cells. RT-qPCR and western
blot analysis revealed that enhanced miR-198 significantly
decreased CDCP1 mRNA and protein levels compared with cells
transfected with control in MCF-7 and MDA-MB-231, respectively
(Fig. 2B-D). Taken together, our
results suggest that CDCP1 is a direct functional target of miR-198
in breast cancer cells.
miR-198 represses cell proliferation
and migration and promotes cell adhesion in breast cancer
cells
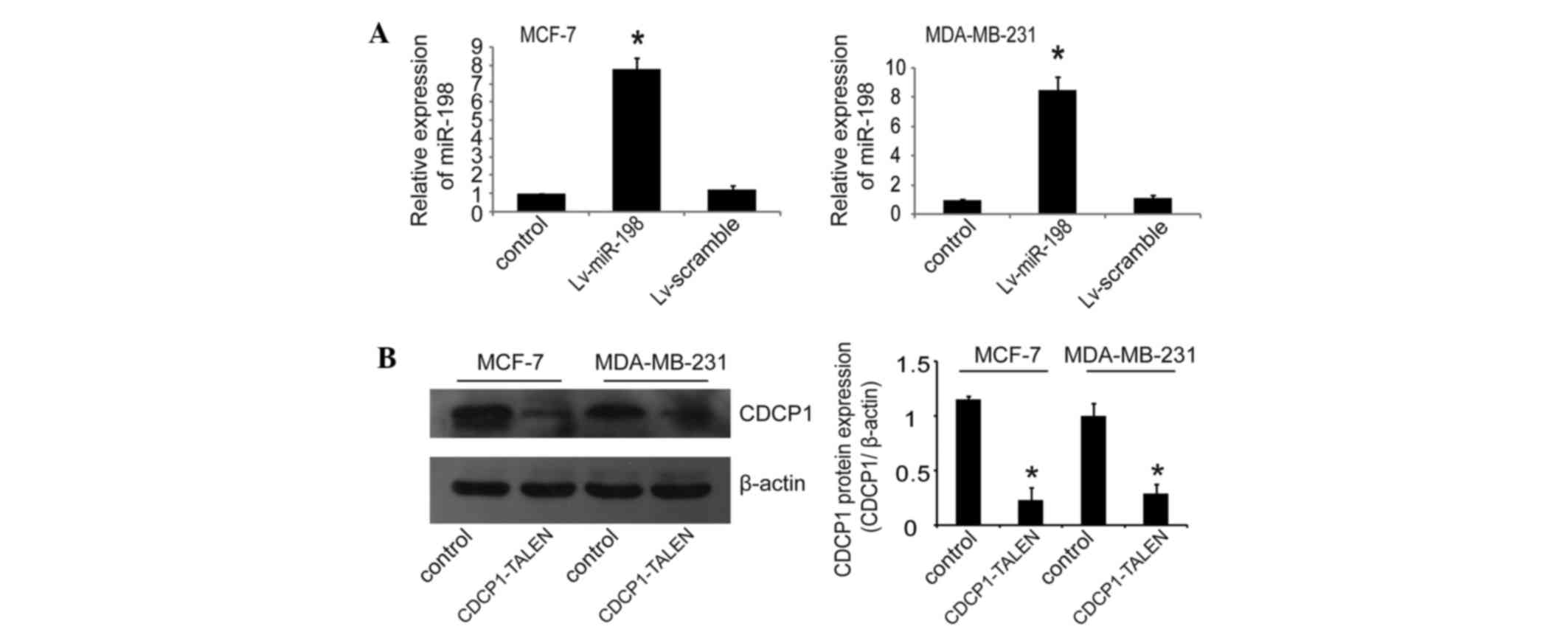
With the understanding that miR-198 is significantly
downregulated in breast cancer tissues, we investigated whether
miR-198 might serve as a tumor suppressor in breast cancer. We
restored miR-198 expression in MCF-7 and MDA-MB-231 cells, which
demonstrated a lower expression of miR-198 in the four selected
breast cancer cell lines, by lentiviral infection with Lv-miR-198
or Lv-scramble lentivirus. RT-qPCR was performed to confirm that
miR-198 was upregulated in MCF-7 and MDA-MB-231 cells following
Lv-miR-198 infection (Fig. 3A). We
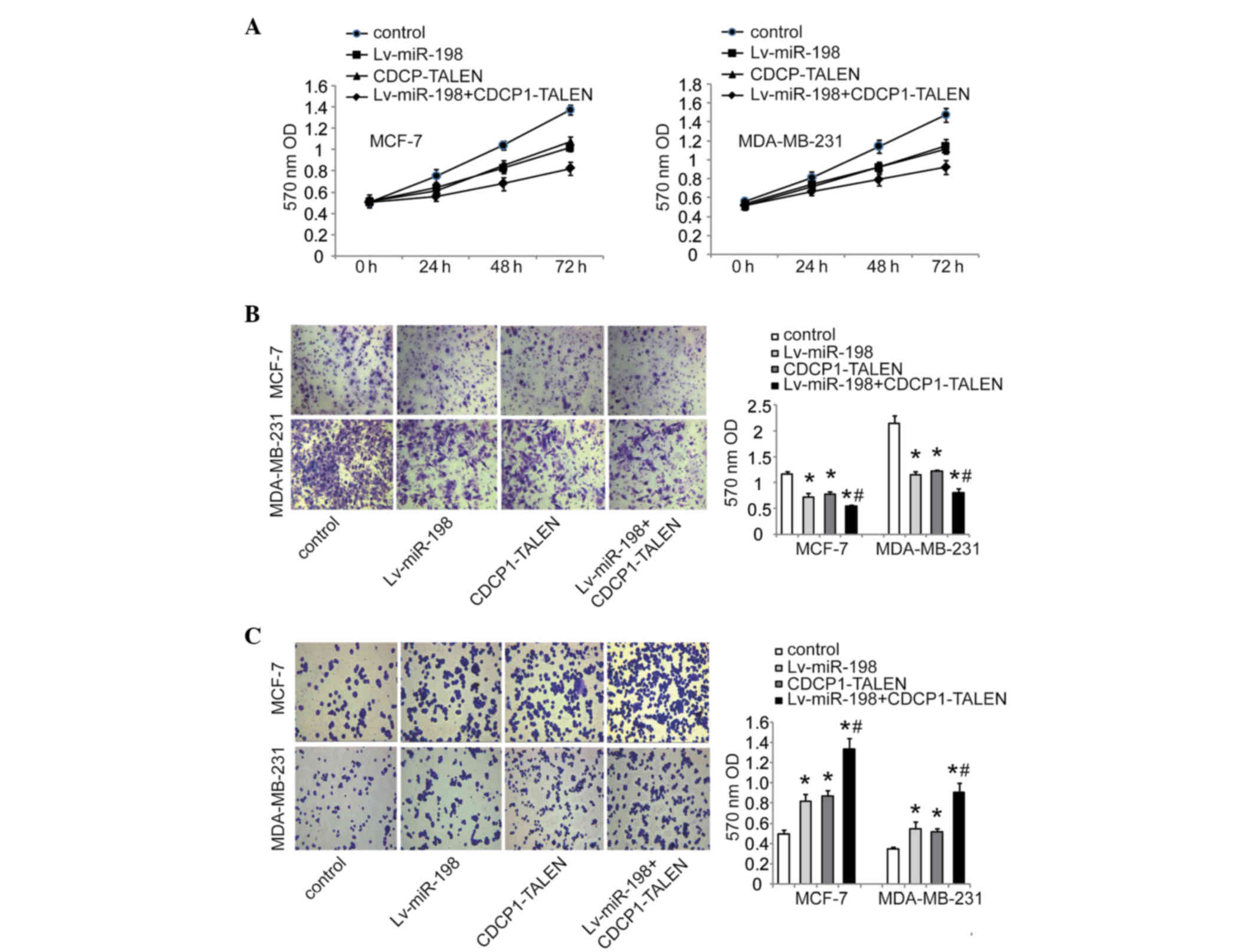
then investigated the effect of miR-198 on cell proliferation,
migration and adhesion, respectively. The MTT assay revealed that
overexpression of miR-198 inhibited the proliferation of MCF-7 and
MDA-MB-231 cells (Fig. 4A). Transwell
assay indicated that the enhanced expression of miR-198 could
significantly inhibit cell migration ability compared with the
control group in MCF-7 and MDA-MB-231 cells (Fig. 4B). Moreover, cell adhesion assays
revealed that miR-198 notably promoted MCF-7 and MDA-MB-231 cell
adhesion (Fig. 4C). Taken together,
our findings suggest that miR-198 may play a suppressive role in
breast cancer cell growth and migration.
Silencing of CDCP1 inhibits cell
proliferation and migration, and promotes cell adhesion in breast
cancer cells
To investigate the precise function of CDCP1 in
breast cancer cells, we next silenced CDCP1 in breast cancer cells.
We knocked out CDCP1 using TALEN technology, which represents a
promising approach for targeted knockout of genes in cultured human
cells (22). Western blot analysis
was performed to confirm TALEN-mediated knockout efficiency in
MCF-7 and MDA-MB-231 cells. As shown in Fig. 3B, the CDCP1 gene was silenced
effectively in CDCP1-TALEN vector-transfected cells. Similar to
miR-198 restoration, silencing CDCP1 by TALEN inhibited cell
proliferation and migration ability (Fig.
4A and B), and promoted cell adhesion (Fig. 4C). We further knocked out CDCP1 in
MCF-7 and MDA-MB-231 cells stably infected with Lv-miR-198. As
expected, a combination of silencing CDCP1 by TALEN and restored
miR-198 had a more enhanced inhibitory effect on proliferation and
migration ability than either silencing CDCP1 by TALEN or restoring
miR-198 alone (Fig. 4A and B). Cell
adhesion assays also revealed that cell adhesion was further
promoted by the combinational treatment (Fig. 4C). Collectively, our results suggest
that CDCP1 is involved in cell growth and migration of breast
cancer cells.
Discussion
In this study, we observed that miR-198 was
downregulated in breast cancer tissues and cell lines compared with
normal cancer tissues and normal cell lines. Then, we demonstrated
that enforced miR-198 inhibited cell proliferation and migration of
breast cancer cells, suggesting that miR-198 may function as a
tumor suppressor in breast cancer metastasis. We also demonstrated
that CDCP1 was upregulated in breast cancer tissues and invasive
cell lines, and was a direct functional target of miR-198. Loss of
function of CDCP1 through TALEN-based knockout suppressed cell
proliferation and migration of breast cancer cells in vitro.
Thus, we reasonably speculate that low expression of miR-198
contributes to CDCP1-mediated cell growth and migration in breast
cancer cells.
miRNAs are frequently dysregulated in various
cancers. Generally, miRNAs affect cancer development through
post-transcriptional regulation of their target genes (23). Thus, the miRNA/target link in certain
cancers may shed light on the molecular mechanism underlying cancer
progression and provide useful potential therapeutic targets for
the clinical treatment of certain cancers. miR-198 was reported to
be located in the 3′UTR of follistatin-like 1 messenger RNA, which
promotes keratinocyte migration, whereas miR-198 expression has the
opposite effect (24). In human
cancers, miR-198 was reported to be downregulated in colorectal
(4), lung (5), pancreatic (6) and hepatocellular carcinoma (8,9), and
generally acts as a tumor suppressor by inhibiting cancer cell
growth and migration. In contrast, high expression of miR-198 was
noted to be associated with a shorter disease-free survival and
overall survival time in pancreatic ductal adenocarcinomas
(7), with poor prognosis in
esophageal cancer (11) and in
high-grade prostate tumors (10).
These data suggest that the roles of miR-198 may vary in different
types of cancer. In the present study, we demonstrated that miR-198
exerted inhibitory effects on the proliferation and migration of
breast cancer cells.
CDCP1 is an integral membrane protein whose
expression is frequently upregulated and positively correlated with
poor prognosis in various types of cancer. CDCP1 was identified as
a protein functionally involved in cancer metastasis in 2003
(13), and was recently shown to
promote migration and peritoneal dissemination of gastric scirrhous
carcinoma (25), and be a unique
target gene of hypoxia-inducible factor 2α involved in the
regulation of renal cancer metastasis (26). In human pancreatic cancers, CDCP1 was
also observed to be a prognostic factor, regulating cell migration
and extracellular matrix degradation (16). In breast cancer, previous studies
suggest that there is a positive correlation between poor prognosis
and high expression of CDCP1 in tumor tissues (13,20). In
the present study, we confirmed that CDCP1 was upregulated in
breast cancer tissues and invasive cell lines. Using MDA-MB-231 and
MCF-7 breast cancer cells as in vitro models, as well as
silencing CDCP1 using TALEN technology, we observed that CDCP1 is
closely associated with cell proliferation, migration and adhesion
in breast cancer cells. Combined with a previous observation that
antibody-mediated CDCP1 degradation significantly inhibited tumor
growth in a mouse xenograft model in vivo (27), we suggest that high expression of
CDCP1 contributes to migration in breast cancer cells and may be
involved in human breast tumorigenesis.
In conclusion, we have established a new
miR-198/CDCP1 link in breast cancer cells, in which the loss of
miR-198 may result in gained expression of CDCP1, which endows
breast cancer cells with improved migration capacity. The
restoration of miR-198 and/or inhibition of CDCP1 expression may be
a promising strategy for breast cancer therapy.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sayed D and Abdellatif M: MicroRNAs in
development and disease. Physiol Rev. 91:827–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang M, Wang J, Kong X, Chen H, Wang Y,
Qin M, Lin Y, Chen H, Xu J, Hong J, et al: miR-198 represses tumor
growth and metastasis in colorectal cancer by targeting fucosyl
transferase 8. Sci Rep. 4:61452014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang J, Zhao H, Xin Y and Fan L:
MicroRNA-198 inhibits proliferation and induces apoptosis of lung
cancer cells via targeting FGFR1. J Cell Biochem. 115:987–995.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marin-Muller C, Li D, Bharadwaj U, Li M,
Chen C, Hodges SE, Fisher WE, Mo Q, Hung MC and Yao Q: A
tumorigenic factor interactome connected through tumor suppressor
microRNA-198 in human pancreatic cancer. Clin Cancer Res.
19:5901–5913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vychytilova-Faltejskova P, Kiss I, Klusova
S, Klusova S, Hlavsa J, Prochazka V, Kala Z, Mazanec J, Hausnerova
J, Kren L, et al: MiR-21, miR-34a, miR-198 and miR-217 as
diagnostic and prognostic biomarkers for chronic pancreatitis and
pancreatic ductal adenocarcinoma. Diagn Pathol. 10:382015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Elfimova N, Sievers E, Eischeid H,
Kwiecinski M, Noetel A, Hunt H, Becker D, Frommolt P, Quasdorff M,
Steffen HM, et al: Control of mitogenic and motogenic pathways by
miR-198, diminishing hepatoma cell growth and migration. Biochim
Biophys Acta. 1833:1190–1198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan S, Li R, Ding K, Lobie PE and Zhu T:
miR-198 inhibits migration and invasion of hepatocellular carcinoma
cells by targeting the HGF/c-MET pathway. FEBS Lett. 585:2229–2234.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Walter BA, Valera VA, Pinto PA and Merino
MJ: Comprehensive microRNA profiling of prostate cancer. J Cancer.
4:350–357. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi B, Yao WJ, Zhao BS, Qin XG, Wang Y,
Wang WJ, Wang TY, Liu SG and Li HC: Involvement of microRNA-198
overexpression in the poor prognosis of esophageal cancer. Asian
Pac J Cancer Prev. 14:5073–5076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Awakura Y, Nakamura E, Takahashi T, Kotani
H, Mikami Y, Kadowaki T, Myoumoto A, Akiyama H, Ito N, Kamoto T, et
al: Microarray-based identification of CUB-domain containing
protein 1 as a potential prognostic marker in conventional renal
cell carcinoma. J Cancer Res Clin Oncol. 134:1363–1369. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hooper JD, Zijlstra A, Aimes RT, Liang H,
Claassen GF, Tarin D, Testa JE and Quigley JP: Subtractive
immunization using highly metastatic human tumor cells identifies
SIMA135/CDCP1, a 135 kDa cell surface phosphorylated glycoprotein
antigen. Oncogene. 22:1783–1794. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ikeda J, Oda T, Inoue M, Uekita T, Sakai
R, Okumura M, Aozasa K and Morii E: Expression of CUB domain
containing protein (CDCP1) is correlated with prognosis and
survival of patients with adenocarcinoma of lung. Cancer Sci.
100:429–433. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu H, Ong SE, Badu-Nkansah K, Schindler
J, White FM and Hynes RO: CUB-domain-containing protein 1 (CDCP1)
activates Src to promote melanoma metastasis. Proc Natl Acad Sci
USA. 108:1379–1384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miyazawa Y, Uekita T, Hiraoka N, Fujii S,
Kosuge T, Kanai Y, Nojima Y and Sakai R: CUB domain-containing
protein 1, a prognostic factor for human pancreatic cancers,
promotes cell migration and extracellular matrix degradation.
Cancer Res. 70:5136–5146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uekita T and Sakai R: Roles of CUB
domain-containing protein 1 signaling in cancer invasion and
metastasis. Cancer Sci. 102:1943–1948. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benes CH, Wu N, Elia AE, Dharia T, Cantley
LC and Soltoff SP: The C2 domain of PKCdelta is a phosphotyrosine
binding domain. Cell. 121:271–280. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uekita T, Jia L, Narisawa-Saito M, Yokota
J, Kiyono T and Sakai R: CUB domain-containing protein 1 is a novel
regulator of anoikis resistance in lung adenocarcinoma. Mol Cell
Biol. 27:7649–7660. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ikeda JI, Morii E, Kimura H, Tomita Y,
Takakuwa T, Hasegawa JI, Kim YK, Miyoshi Y, Noguchi S, Nishida T
and Aozasa K: Epigenetic regulation of the expression of the novel
stem cell marker CDCP1 in cancer cells. J Pathol. 210:75–84. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seidel J, Kunc K, Possinger K, Jehn C and
Lüftner D: Effect of the tyrosine kinase inhibitor lapatinib on
CUB-domain containing protein (CDCP1)-mediated breast cancer cell
survival and migration. Biochem Biophys Res Commun. 414:226–232.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cermak T, Doyle EL, Christian M, Wang L,
Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ and Voytas
DF: Efficient design and assembly of custom TALEN and other TAL
effector-based constructs for DNA targeting. Nucleic Acids Res.
39:e822011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sundaram GM, Common JE, Gopal FE, Srikanta
S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB and Sampath P:
‘See-saw’ expression of microRNA-198 and FSTL1 from a single
transcript in wound healing. Nature. 495:103–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uekita T, Tanaka M, Takigahira M, Miyazawa
Y, Nakanishi Y, Kanai Y, Yanagihara K and Sakai R:
CUB-domain-containing protein 1 regulates peritoneal dissemination
of gastric scirrhous carcinoma. Am J Pathol. 172:1729–1739. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Emerling BM, Benes CH, Poulogiannis G,
Bell EL, Courtney K, Liu H, Choo-Wing R, Bellinger G, Tsukazawa KS,
Brown V, et al: Identification of CDCP1 as a hypoxia-inducible
factor 2α (HIF-2α) target gene that is associated with survival in
clear cell renal cell carcinoma patients. Proc Natl Acad Sci USA.
110:3483–3488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kollmorgen G, Niederfellner G, Lifke A,
Spohn GJ, Rieder N, Harring SV, Bauss F, Burtscher H, Lammers R and
Bossenmaier B: Antibody mediated CDCP1 degradation as mode of
action for cancer targeted therapy. Mol Oncol. 7:1142–1151. 2013.
View Article : Google Scholar : PubMed/NCBI
|