Introduction
Esophageal cancer is a common malignant tumor that
represents the sixth leading cause of cancer-associated mortality,
worldwide (1). Esophageal squamous
cell carcinoma (ESCC) accounts for >90% of all esophageal cancer
cases (2). ESCC is a potentially
fatal malignancy that arises from esophageal epithelial cells, that
is common in China, with a high global incidence (3). Although diagnosis and treatment of ESCC
have improved in recent years, ESCC is frequently diagnosed in
patients with advanced stage disease and subsequently, tumors are
unresectable (4,5). The identification of early diagnostic
and prognostic evaluation markers are required to improve diagnosis
and prognosis of ESCC.
The human chemokine-like factor (CKLF)-like MARVEL
transmembrane domain-containing (CMTM) family is a novel gene
family, consisting of nine genes, CKLF and CMTM1-8 (6,7). CMTM
proteins exhibit critical functions in the immune system, male
reproductive system and tumorigenesis (8–16). CMTM3,
which is a member of the chemokine-like factor gene superfamily, is
similar to the chemokine and transmembrane 4 superfamily (TM4SF) of
signaling molecules in that they both have 4 transmembrane regions
(7). The TM4SF family contains a
number of tumor-associated genes, including, TM4SF/plasmolipin, MAL
and BENE, which exhibit strong tumor-suppressive functions in
esophageal and prostate cancer (17,18).
CMTM3, which is located at 16q22.1, has frequently been identified
in a number of carcinomas, including prostate tumors, oral squamous
cell carcinoma and testicular cancer (11,19,20).
Therefore, the present study hypothesized that CMTM3 may act as a
tumor suppressor gene (TSG). A previous study demonstrated that
CMTM3 was downregulated in 7/18 esophageal cell lines (21). However, the association between CMTM3
expression and the prognosis and clinicopathological features of
ESCC patients remains unclear.
The aim of the present study was to investigate
association between CMTM3 expression and prognosis in ESCC.
Quantitative reverse transcription-polymerase chain reaction
(qRT-PCR) and western blotting were used to analyze CMTM3 mRNA and
protein expression levels in 40 ESCC tissues and 40 adjacent
non-tumor tissues. Immunohistochemistry was performed to
investigate the clinical relevance of CMTM3 expression in an
additional 110 ESCC tissues and 36 adjacent non-tumor tissues.
Materials and methods
Human samples
A total of 40 paired ESCC and normal esophageal
tissues were obtained from ESCC patients who underwent complete
esophageal cancer resection at The First Affiliated Hospital of
China Medical University (Shenyang, China) between April 2014 and
August 2014. Patients who underwent palliative surgery were not
included. The mean age of the ESCC patients was 57.71 years (range,
44–71 years). The fresh tissues were immediately frozen in liquid
nitrogen and stored at −80°C prior to RNA and protein extraction.
To investigate the association between CMTM3 expression and the
clinicopathological characteristics and prognosis of ESCC patients,
an additional 110 archived paraffin-embedded specimens and 36
adjacent non-tumor paraffin-embedded tissues, obtained from ESCC
patients who underwent complete esophageal cancer resection at the
First Affiliated Hospital of China Medical University between May
2008 and April 2010, were included in this study for
immunohistochemical (IHC) analysis. None of the 110 patients
received neoadjuvant therapy prior to surgery. The mean age of the
ESCC patients was 60.24 years (range, 43–80 years). The
differentiation grade, tumor-node-metastasis (TNM) stage and lymph
node status were classified according to the International Union
Against Cancer/American Joint Committee on Cancer TNM
classification (7th edition) (22).
Clinicopathological data, which included age, gender, tumor
location, TNM stage, differentiation and lymph node metastasis, was
obtained from medical records (Table
I). Overall survival was determined from the date of surgery to
the time of death, estimated by the Kaplan Meier method. The
present study was approved by the Speciality Committee on Ethics of
Biomedicine Research of The First Affiliated Hospital of China
Medical University. Written informed consent was obtained from all
patients.
 | Table I.Associations between CMTM3 expression
and clinicopathological features of 110 ESCC patients. |
Table I.
Associations between CMTM3 expression
and clinicopathological features of 110 ESCC patients.
|
|
| CMTM3 expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameters | Patients, n | Low | High | P-value |
|---|
| Age, years |
|
|
| 0.280 |
| ≤60 | 48 | 37 | 11 |
|
|
>60 | 62 | 42 | 20 |
|
| Gender |
|
|
| 0.275 |
| Male | 97 | 68 | 29 |
|
|
Female | 13 | 11 | 2 |
|
| Location |
|
|
| 0.357 |
| Ut | 26 | 19 | 7 |
|
| Mt | 32 | 20 | 12 |
| Lt | 52 | 40 | 12 |
| Pathological
grade |
|
|
| 0.730 |
|
Moderately/poorly-differentiated | 61 | 43 | 18 |
|
Well-differentiated | 49 | 36 | 13 |
| Lymph node
metastasis |
|
|
| 0.002 |
|
Negative | 56 | 33 | 23 |
|
Positive | 54 | 46 | 8 |
| Clinical stage |
|
|
| 0.001 |
| I+II | 58 | 33 | 25 |
|
III+IV | 52 | 46 | 6 |
qRT-PCR
Total RNA (2 µg) was extracted from tissues using
TaKaRa RNAiso Reagent (Takara Bio, Inc., Otsu, Japan) according to
manufacturer's instructions. The quantity and quality of the
Extracted RNA were analyzed by spectrophotometer at 260 nm (ND1000;
Nanodrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). RNA
(2 µg) was reverse-transcribed using PrimeScript RT Reagent Kit
with gDNA Eraser (Takara Bio, Inc.) to synthesize cDNA. qRT-PCR was
performed using SYBR® Premix Ex Taq™ II (Takara Bio,
Inc.) and the Corbet Rotor-Gene 3000 thermocycler (Rotor-Gene 3000;
Corbett Research, Mortlake, Australia). Glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) served as an internal control. The following
primers were used: Sense, 5′-GCTTGTGCTGGCCCATGATG-3′ and antisense,
5′-TGTGGGCTGTGGTCTCATCT-3′ for CMTM3; sense,
5′-CTCCTCCTGTTCGACAGTCAGC-3′ and antisense,
5′-CCCAATACGACCAAATCCGTT-3′ for GAPDH. PCR amplification was
performed under the following conditions: 95°C for 1 min, followed
by 35 cycles at 95°C for 15 sec and 60°C for 1 min. Experiments
were performed in triplicate. CMTM3 mRNA expression was quantified
relative to that of GAPDH according to the comparative threshold
cycle (2−ΔΔCq) method (23).
Western blot analysis
A total of 40 paired ESCC and non-tumor tissue
samples were sectioned, homogenized and lysed in RIPA lysis buffer
supplemented with 1% (v/v) protease inhibitor cocktail and
phenylmethylsulfonyl fluoride (all Beyotime Institute of Biology,
Haimen, China). Protein concentrations were determined using the
BCA method with an Enhanced BCA Protein Assay kit (P0009; Beyotime
Institute of Biotechnology) and a microplate reader (BioTek,
Winooski, VT, USA) according to the manufacturer's instructions.
Proteins (50 µg/lane) were separated by 12% SDS-PAGE and
transferred to polyvinylidine difluoride filter membranes (EMD
Millipore, Billerica, MA, USA) in a wet transfer system (Bio-Rad,
Berkeley, CA, USA) at 70 V. Next, Tris-buffered saline with
Tween-20 (TBST) containing 5% nonfat milk was used to block the
membrane for 1 h at room temperature, followed by incubation with
CMTM3 (cat. no. ab198016; 1:1,000; Abcam, Cambridge, UK) and GAPDH
(cat. no. ab181602; 1:5,000; Abcam) primary antibodies at 4°C
overnight. After washing with TBST, the membranes were incubated
with horseradish peroxidase-conjugated goat anti-rabbit secondary
antibodies (cat. no. sc-2004; 1:10,000; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) at room temperature for 1 h. Immunopositive
bands were visualized using enhanced chemiluminescence buffer
(Beyotime Institute of Biotechnology) and band intensities were
quantified using MF-ChemiBIS 2.0 (DNR Bio-Imaging Systems Ltd.,
Jerusalem, Israel) and Quantity One software (version 4.62;
Bio-Rad).
IHC analysis
A total of 110 paraffin-embedded biopsy samples were
cut into 4-µm sections, dewaxed, rehydrated through decreasing
concentrations of ethanol citrate buffer and heated in 10 mmol/l
sodium citrate buffer (pH 6.0) for antigen retrieval (high-pressure
heat method for 5 min). After washing in phosphate-buffered saline
(Beyotime Institute of Biotechnology), all tissue slides were
blocked with 10% normal goat serum for 30 min and incubated with
CMTM3 primary antibody (cat. no. ab198016; 1:100; Abcam) in a
humidified chamber at 4°C over night. Membranes were then incubated
with horseradish peroxidase-conjugated IgG secondary antibodies
(1:1; PV-9000; Beijing Zhongshan Golden Bridge Biotechnology Co.,
Ltd., Beijing, China) at 37°C for 10 min. Next, the sections were
incubated with DAB solution (Beyotime Institute of Biotechnology)
for 3 min and counterstained with hematoxylin (Beyotime Institute
of Biotechnology). The expression of CMTM3 was determined by two
independent pathologists blinded to the clinical data. Briefly,
staining intensity was scored on a 4-tiered scale (0, no
immunostaining; 1, light-brown color; 2, medium-brown color; 3,
brown color). The percentage of positively stained cells per field
was defined as: 0, ≤5%; 1, 5–25%; 2, 26–50%; 3, >50%. The score
for each field was the product of the percentage and intensity
scores and the final score for CMTM3 expression in each case was
the mean score of five fields: -, 0 points; +, 1–2 points; ++, 3–5
points; +++, 6–9 points. For analysis, the patients were divided
into CMTM3 ‘high expression’ (++ and +++) and ‘low expression’ (+
and-) groups. Discrepancies were resolved by discussion between the
pathologists.
Statistical analysis
Data are presented as the mean ± standard deviation.
All statistical analysis was performed using SPSS 21.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Comparison of CMTM3
expression between ESCC tissues and the corresponding adjacent
non-tumor tissues was performed using the paired Student's t-test.
χ2 test was used to analyze the correlation between
CMTM3 expression and patient clinicopathological features. The
Kaplan Meier method was used to plot survival curves and results
were analyzed using the log-rank test. Cox proportional hazards
regression model was used for univariate and multivariate survival
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
CMTM3 mRNA expression levels are
decreased in ESCC tissues compared with adjacent non-tumor
tissues
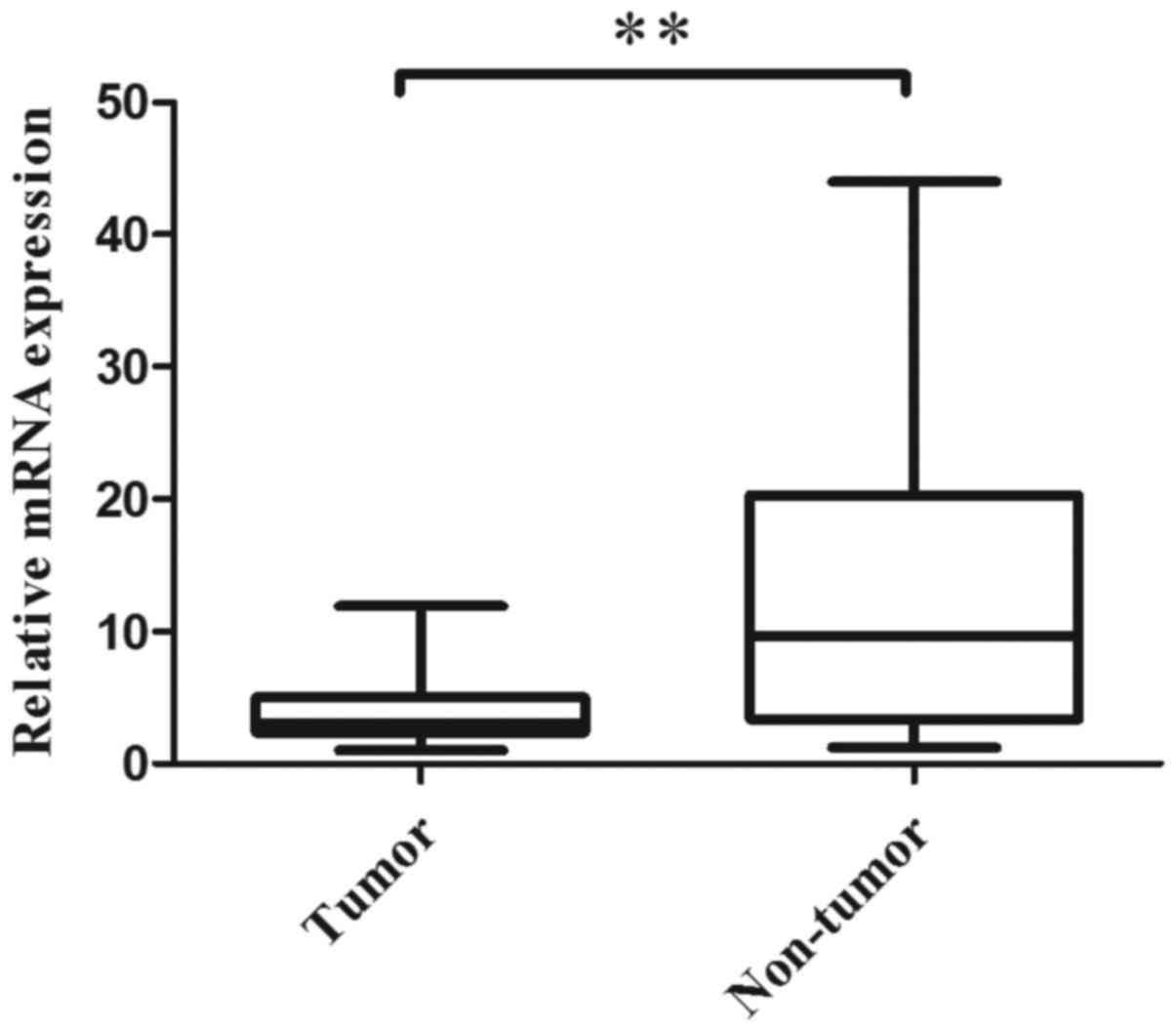
qRT-PCR was performed to evaluate the CMTM3 mRNA
expression levels in 40 paired normal and ESCC tumor tissues. The
results revealed that 82.5% (33/40) of tumor tissues expressed
lower CMTM3 mRNA levels than their adjacent non-tumor tissues
(P<0.001; Fig. 1). The mean
relative expression levels of CMTM3 mRNA in the tumor and non-tumor
tissues were 4.01 and 13.09, respectively. The results indicated
that CMTM3 mRNA expression is significantly decreased in tumor
tissues when compared with the adjacent non-tumor tissues.
CMTM3 protein expression levels are
decreased in ESCC tissues compared with adjacent non-tumor
tissues
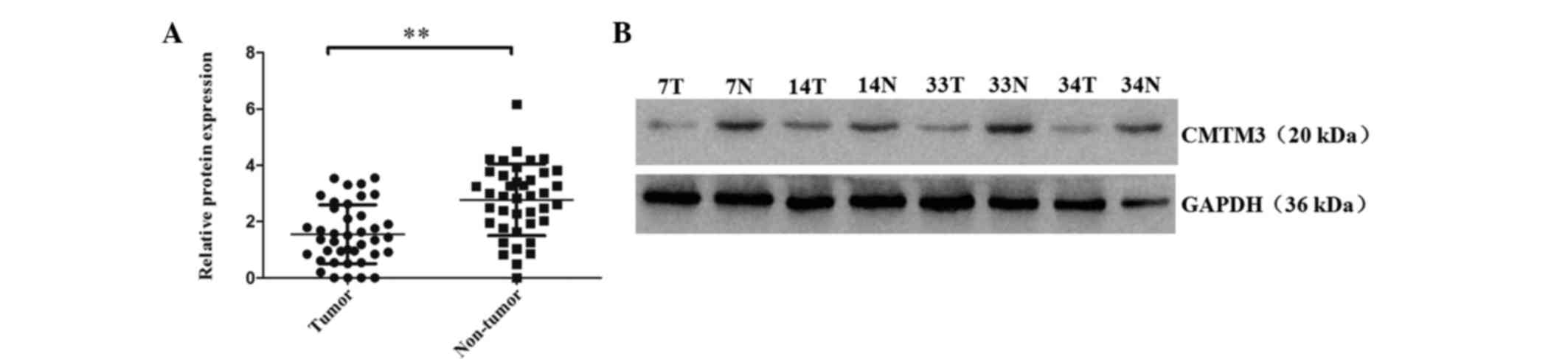
CMTM3 protein levels in 40 paired of ESCC tumor and
adjacent non-tumor tissues were analyzed by western blot analysis.
The results revealed that 75% (30/40) of tumor tissues expressed
lower CMTM3 protein expression levels than their adjacent non-tumor
tissues (P<0.001; Fig. 2). The
mean relative expression levels of CMTM3 protein in the tumor and
non-tumor tissues were 1.55 and 2.77, respectively. These results
indicated that CMTM3 protein expression is significantly decreased
in ESCC tumor tissue when compared with the paired adjacent
non-tumor tissues. Furthermore, low CMTM3 expression may be
important in the tumorigenesis of ESCC.
CMTM3 protein expression is associated
with lymph node metastasis and clinical stage in ESCC tissues
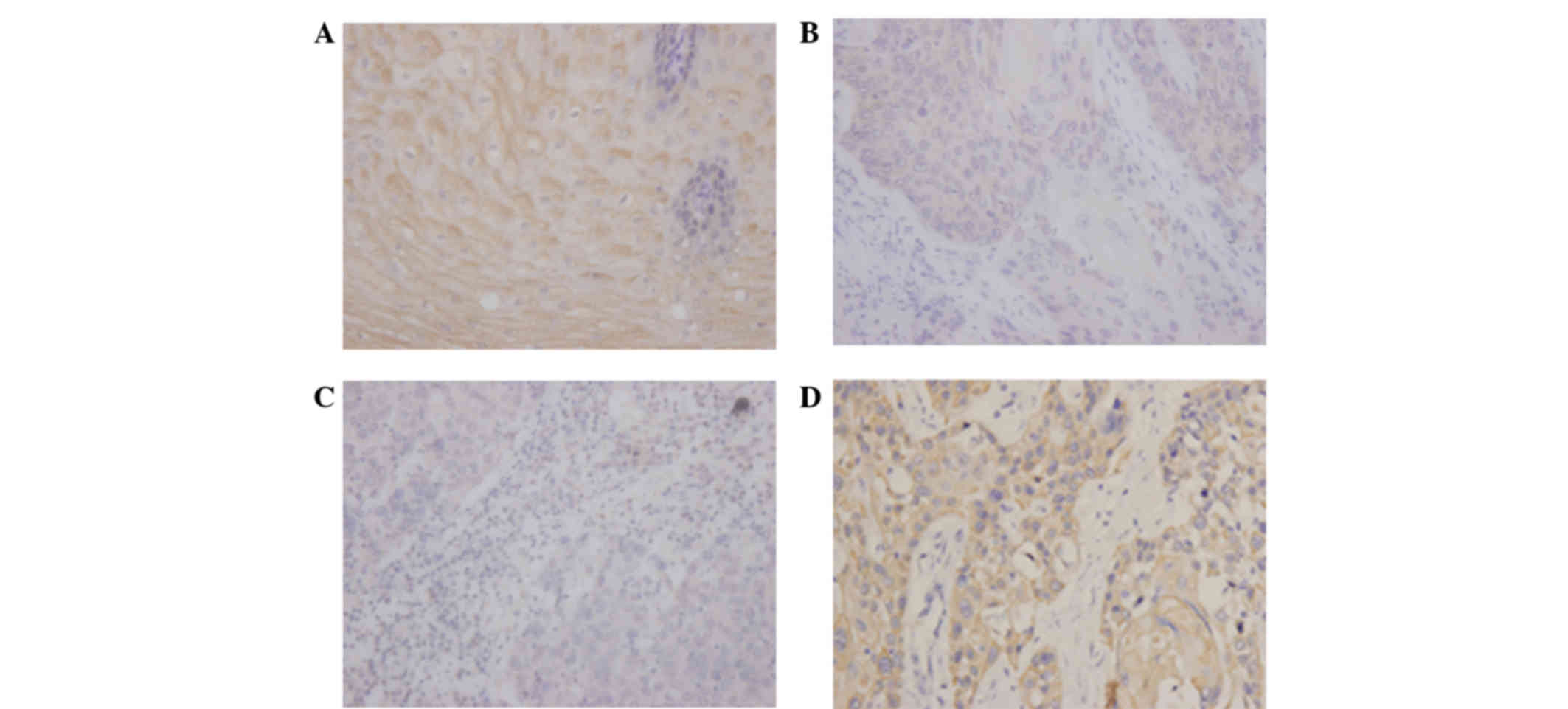
To investigate the association between CMTM3 protein
expression and clinicopathological parameters, a total of 110
tissues were obtained from 110 ESCC patients. According to the
level of CMTM3 expression in tumor tissues as determined by
immunohistochemistry analysis (Fig.
3), the ESCC patients were divided into low and high expression
groups. No significant associations were identified between CMTM3
expression and age (P=0.280), gender (P=0.275), tumor location
(P=0.357) or pathological grade (P=0.730), however, CMTM3
expression was associated with lymph node metastasis (P=0.002) and
clinical stage (P<0.001) in ESCC tissues (Table I).
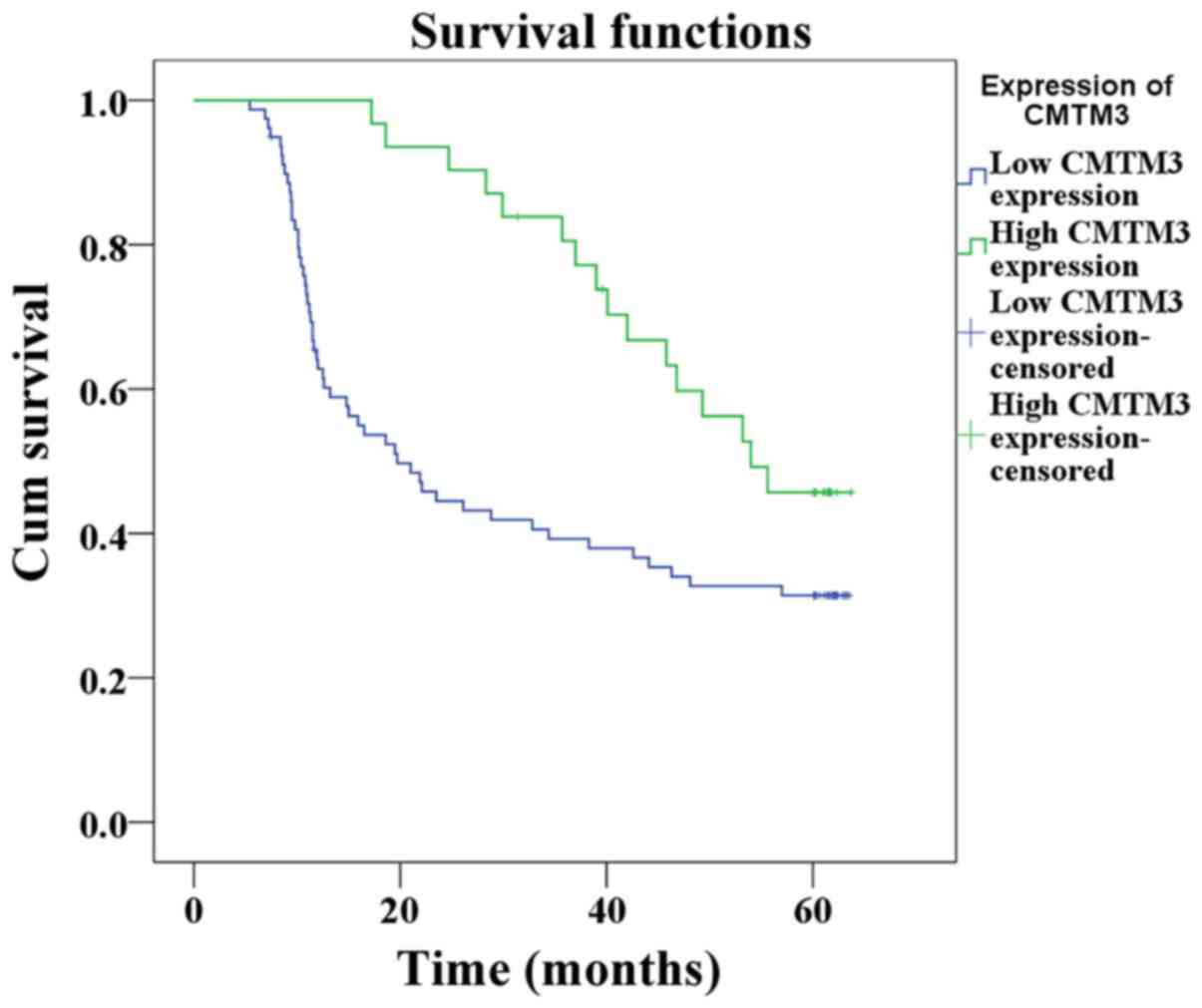
Survival time of patients exhibiting
low CMTM3 expression is significantly shorter than patients
exhibiting high CMTM3 expression in ESCC
The estimated survival time was calculated according
to the Kaplan-Meier method. The median duration of follow up was
35.05 months (range, 5–64 months). Patients that succumbed as a
result of postoperative complications were not included in the
study. The results revealed that the survival time of the low CMTM3
expression group was significantly shorter than that of the high
CMTM3 expression group (32.9 vs. 48.4 %P=0.010; Fig. 4).
Cox regression analysis was carried out to assess
whether CMTM3 is an independent prognostic factor for survival in
ESCC. Univariate analysis revealed that patient overall survival
was significantly associated with pathological grade (P=0.012),
lymph node metastasis (P<0.001), clinical stage (P<0.001) and
CMTM3 expression (P=0.012). Multivariate analysis revealed that
CMTM3 expression (P=0.036), pathological grade (P=0.001), clinical
stage (P<0.001) and lymph node metastasis (P=0.001) were
independent prognostic factors for overall survival in ESCC
patients (Table II).
 | Table II.Univariate and multivariate analysis
of overall survival in 110 ESCC patients. |
Table II.
Univariate and multivariate analysis
of overall survival in 110 ESCC patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age, years | 0.726 |
| 0.897 |
|
≤60 |
| 1 |
|
|
>60 |
| 0.967
(0.580–1.580) |
|
| Gender | 0.413 |
| 0.287 |
|
Male |
| 1 |
|
|
Female |
| 0.727
(0.353–1.353) |
|
| Location |
|
| 0.903 |
| Ut | 0.901 | 1 |
|
| Mt | 0.918 | 1.065
(0.583–1.583) | 0.838 |
| Lt | 0.704 | 1.059
(0.579–1.579) | 0.853 |
| Pathological
grade | 0.012 |
| 0.001 |
|
Well-differeniated |
| 1 |
|
|
Moderately/poorly-differentiated |
| 2.594
(1.516–4.516) |
|
| Lymph node
metastasis | <0.001 |
| 0.001 |
|
Negative |
| 1 |
|
|
Positive Clinical |
| 2.863
(1.574–5.574) |
|
| Clinical stage | <0.001 |
| <0.001 |
|
IA-IIB |
| 1 |
|
|
IIIA-IV |
| 3.710
(1.994–6.994) |
|
| CMTM3
expression | 0.012 |
| 0.036 |
|
High |
| 1 |
|
|
Low |
| 1.961
(1.046–3.046) |
|
Discussion
CMTM3, which is a member of the CMTM family that was
first identified by Han et al (7) in 2003. CMTM3 is located at 16q22.1, an
important tumor suppressor locus that is associated with the
pathogenesis of multiple carcinomas (24–26).
Previous studies have demonstrated that CMTM3 is a candidate tumor
suppressor gene in a number of tumors, such as renal cell carcinoma
(10), testicular cancer (11), gastric cancer (13) and oral squamous cell carcinoma
(19). Previously CMTM3 was
demonstrated to be silenced or downregulated in 7/18 esophageal
cell lines (21), however, CMTM3
expression in ESCC and its association with prognosis remains
unknown.
In the present study, qRT-PCR revealed that 82.5%
(33/40) of ESCC tissues expressed lower levels of CMTM3 mRNA
expression compared with adjacent non-tumor tissues. Consistent
with these results, western blot analysis revealed that 75% (30/40)
of ESCC tissues expressed lower levels of CMTM3 protein compared
with adjacent non-tumor tissues. Furthermore, IHC analysis was
performed to analyze associations between CMTM3 expression and
clinicopathological features in 110 ESCC patients. No significant
association was identified between CMTM3 expression and patient age
(P=0.280), gender (P=0.275), tumor location (P=0.357) or
pathological grade (P=0.730), however, CMTM3 expression was
significantly associated with lymph node metastasis (P=0.002) and
clinical stage (P<0.001) in ESCC tissues. IHC analysis also
revealed that of the 110 ESCC samples, 79 cases (71.82%) exhibited
low CMTM3 expression and 31 cases (28.18%) exhibited high CMTM3
expression. Of the 36 adjacent non-tumor tissues, 27 cases (75%)
exhibited high CMTM3 expression and 9 cases (25%) exhibited low
CMTM3 expression. These results indicate that CMTM3 may be involved
in the progression of ESCC and may act as a tumor suppressor in
ESCC. In the present study, the correlation between CMTM3
expression and patient prognosis was evaluated in ESCC patients.
Kaplan-Meier survival analysis revealed that the overall survival
time of patients with low CMTM3 expression was significantly
shorter than patients with high CMTM3 expression (P=0.010). Cox
multivariate analysis indicated that CMTM3 protein expression was
an independent prognostic predictor for ESCC after resection.
CpG methylation resulting in the loss of TSG
functions is a major epigenetic alteration that leads to tumor
development and progression (27).
The CMTM3 promoter contains a typical CpG island consisting of 53
CpG sites, which is methylated by the addition of a methyl group
via DNA methyltransferase enzymes (21). However, a previous study reported that
CMTM3 was methylated in 3% of esophageal carcinomas (21). Consistent with these results,
methylation of the promoter region of CMTM3 is not observed in
renal cell carcinoma (10). Thus, it
may be hypothesized that unlike the aberrant methylation observed
in tumors such as oral squamous cell carcinoma (20), hepatocellular carcinoma (28) and gastric cancer (21), in ESCC low expression of CMTM3 may be
associated with other genetic or epigenetic mechanisms. The results
of the present indicated that CMTM3 is expressed in a number of
ESCC tissues, however, its function in ESCC cell lines remains
unclear. Thus, further studies are required to increase
understanding with regard to the function of CMTM3 in ESCC
cells.
In conclusion, in the present study CMTM3 expression
was significantly decreased in ESCC tissue compared with adjacent
non-tumor tissue. Furthermore, this study is the first to indicate
that CMTM3 protein expression in resected tumors is an effective
prognostic biomarker for ESCC.
Acknowledgements
This study was supported by the National Nature
Science Foundation of China (grant no. 81201890) and the Natural
Science Foundation of Liaoning Province (grant no. 2013021002).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hongo M, Nagasaki Y and Shoji T:
Epidemiology of esophageal cancer: Orient to Occident. Effects of
chronology, geography and ethnicity. J Gastroenterol Hepatol.
24:729–735. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun ZG, Wang Z, Liu XY and Liu FY: Mucin 1
and vascular endothelial growth factor C expression correlates with
lymph node metastatic recurrence in patients with N0 esophageal
cancer after Ivor-Lewis esophagectomy. World J Surg. 35:70–77.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thallinger CM, Kiesewetter B, Raderer M
and Hejna M: Pre- and postoperative treatment modalities for
esophageal squamous cell carcinoma. Anticancer Res. 32:4609–4627.
2012.PubMed/NCBI
|
|
5
|
Kranzfelder M, Büchler P and Friess H:
Surgery within multimodal therapy concepts for esophageal squamous
cell carcinoma (ESCC): The MRI approach and review of the
literature. Adv Med Sci. 54:158–169. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han W, Lou Y, Tang J, Zhang Y, Chen Y, Li
Y, Gu W, Huang J, Gui L, Tang Y, et al: Molecular cloning and
characterization of chemokine-like factor 1 (CKLF1), a novel human
cytokine with unique structure and potential chemotactic activity.
Biochem J. 357:127–135. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han W, Ding P, Xu M, Wang L, Rui M, Shi S,
Liu Y, Zheng Y, Chen Y, Yang T and Ma D: Identification of eight
genes encoding chemokine-like factor superfamily members 1–8
(CKLFSF1-8) by in silico cloning and experimental validation.
Genomics. 81:609–617. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Imamura Y, Katahira T and Kitamura D:
Identification and characterization of a novel BASH N
terminus-associated protein, BNAS2. J Biol Chem. 279:26425–26432.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Li T, Qiu X, Mo X, Zhang Y, Song
Q, Ma D and Han W: CMTM3 can affect the transcription activity of
androgen receptor and inhibit the expression level of PSA in LNCaP
cells. Biochem Biophys Res Commun. 371:54–58. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie J, Yuan Y, Liu Z, Xiao Y, Zhang X, Qin
C, Sheng Z, Xu T and Wang X: CMTM3 is frequently reduced in clear
cell renal cell carcinoma and exhibits tumor suppressor activities.
Clin Transl Oncol. 16:402–409. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Z, Xie J, Wu J, Li W, Nie L, Sun X,
Tang A, Li X, Liu R, Mei H, et al: CMTM3 inhibits human testicular
cancer cell growth through inducing cell-cycle arrest and
apoptosis. PLoS One. 9:e889652014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dworakowska D, Wlodek E, Leontiou CA,
Igreja S, Cakir M, Teng M, Prodromou N, Góth MI, Grozinsky-Glasberg
S, Gueorguiev M, et al: Activation of RAF/MEK/ERK and PI3K/AKT/mTOR
pathways in pituitary adenomas and their effects on downstream
effectors. Endocr Relat Cancer. 16:1329–1338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Su Y, Lin Y, Zhang L, Liu B, Yuan W, Mo X,
Wang X, Li H, Xing X, Cheng X, et al: CMTM3 inhibits cell migration
and invasion and correlates with favorable prognosis in gastric
cancer. Cancer Sci. 105:26–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shao L, Cui Y, Li H, Liu Y, Zhao H, Wang
Y, Zhang Y, Ng KM, Han W, Ma D and Tao Q: CMTM5 exhibits tumor
suppressor activities and is frequently silenced by methylation in
carcinoma cell lines. Clin Cancer Res. 13:5756–5762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chowdhury MH, Nagai A, Terashima M, Sheikh
A, Murakawa Y, Kobayashi S and Yamaguchi S: Chemokine-like factor
expression in the idiopathic inflammatory myopathies. Acta Neurol
Scand. 118:106–114. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li D, Jin C, Yin C, Zhang Y, Pang B, Tian
L, Han W, Ma D and Wang Y: An alternative splice form of CMTM8
induces apoptosis. Int J Biochem Cell Biol. 39:2107–2119. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mimori K, Shiraishi T, Mashino K, Sonoda
H, Yamashita K, Yoshinaga K, Masuda T, Utsunomiya T, Alonso MA,
Inoue H and Mori M: MAL gene expression in esophageal cancer
suppresses motility, invasion and tumorigenicity and enhances
apoptosis through the Fas pathway. Oncogene. 22:3463–3471. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang XA, He B, Zhou B and Liu L:
Requirement of the p130CAS-Crk coupling for metastasis suppressor
KAI1/CD82-mediated inhibition of cell migration. J Biol Chem.
278:27319–27328. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu F, Yuan W, Wang X, Sheng Z, Yuan Y, Qin
C, He C and Xu T: CMTM3 is reduced in prostate cancer and inhibits
migration, invasion and growth of LNCaP cells. Clin Transl Oncol.
17:632–639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang H, Zhang J, Nan X, Li X, Qu J, Hong
Y, Sun L, Chen Y and Li T: CMTM3 inhibits cell growth and migration
and predicts favorable survival in oral squamous cell carcinoma.
Tumour Biol. 36:7849–7858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Li J, Cui Y, Li T, Ng KM, Geng H,
Li H, Shu XS, Li H, Liu W, et al: CMTM3, located at the critical
tumor suppressor locus 16q22.1, is silenced by CpG methylation in
carcinomas and inhibits tumor cell growth through inducing
apoptosis. Cancer Res. 69:5194–5201. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caldeira JR, Prando EC, Quevedo FC, Neto
FA, Rainho CA and Rogatto SR: CDH1 promoter hypermethylation and
E-cadherin protein expression in infiltrating breast cancer. BMC
Cancer. 6:482006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chan KY, Lai PB, Squire JA, Beheshti B,
Wong NL, Sy SM and Wong N: Positional expression profiling
indicates candidate genes in deletion hotspots of hepatocellular
carcinoma. Mod Pathol. 19:1546–1554. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li C, Berx G, Larsson C, Auer G, Aspenblad
U, Pan Y, Sundelin B, Ekman P, Nordenskjöld M, van Roy F and
Bergerheim US: Distinct deleted regions on chromosome segment
16q23-24 associated with metastases in prostate cancer. Genes
Chromosomes Cancer. 24:175–182. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
28
|
Zhao W, Xu Y, Xu J, Wu D, Zhao B, Yin Z
and Wang X: Subsets of myeloid-derived suppressor cells in
hepatocellular carcinoma express chemokines and chemokine receptors
differentially. Int Immunopharmacol. 26:314–321. 2015. View Article : Google Scholar : PubMed/NCBI
|